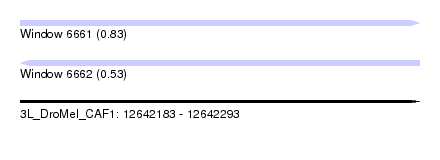
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,642,183 – 12,642,293 |
| Length | 110 |
| Max. P | 0.825458 |

| Location | 12,642,183 – 12,642,293 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.42 |
| Mean single sequence MFE | -34.42 |
| Consensus MFE | -23.37 |
| Energy contribution | -24.73 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
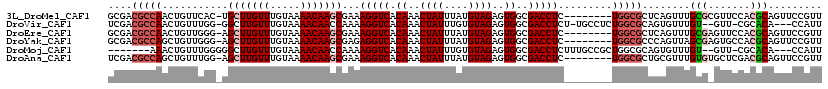
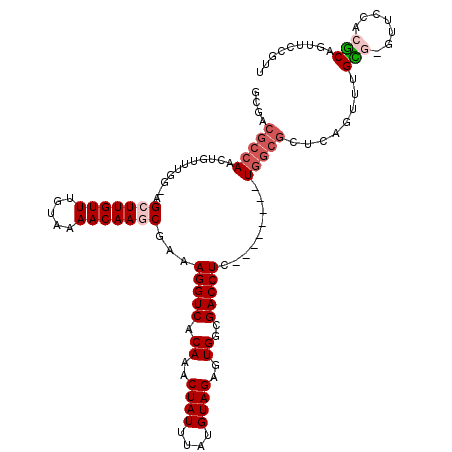
>3L_DroMel_CAF1 12642183 110 + 23771897 AACGGAACUGCGUGGAACGCGCAAACUGAGCGCCA--------GAGGUCGCCACUCUACAUAAAUAGUUUGUGACCUUUCGCUUGUUUUACAAACAAGCA-GUGAACAGUUGGCGUCGC ..(((...((((((...))))))..))).((((((--------((((((((....(((......)))...))))))))..(((((((.....))))))).-.........))))))... ( -37.20) >DroVir_CAF1 6430 111 + 1 AAUGG---UGUGCG-AAC--ACAAACACUGCGCCAGAGGCA-AGAGGUCGCCACUCUACACAAAUAGUUUGUGACCUUUUGGUUGUUUUACAAACAAGCC-CCAAACAGUUGGCGUCGA ....(---(((...-.))--)).....(.(((((((..(((-(((((((((....(((......)))...))))))))))).(((((.....)))))...-.....)..))))))).). ( -33.00) >DroEre_CAF1 6505 110 + 1 AACGGAACUGCGUGGAACUCGCAAACUGAGCGCCA--------GAGGUCGCCACUCUACAUAAAUAGUUUGUGACCUUUCGCUUGUUUUACAAACAAGCU-CCCAACAGUUGGCGUCGC ..(((...((((.......))))..))).((((((--------((((((((....(((......)))...))))))))..(((((((.....))))))).-.........))))))... ( -34.10) >DroYak_CAF1 5915 110 + 1 AACGGAACUGCGUGGCACUCGCUAACUGGGCGCCA--------GAGGUCGCCACUCUACAUAAAUAGUUUGUGACCUCUCGCUUGUUUUACAAACAAGCU-CCCAACAGCUGGCGUCGC ..(((....(((.......)))...)))(((((((--------((((((((....(((......)))...))))))))..(((((((.....))))))).-.........))))))).. ( -39.00) >DroMoj_CAF1 6989 106 + 1 AAUGG---UGUGCG-AAC--ACAAACACUGCGCCAGCGGCAAAGAGGUCGCCACUCUACACAAAUAGUUUGUGACCUUUUGGUUGUUUUACAAACAAGCCCCCAAACAGUUU------- ....(---(((...-.))--))((((...((....))(((.((((((((((....(((......)))...))))))))))..(((((.....))))))))........))))------- ( -26.50) >DroAna_CAF1 7049 110 + 1 AACGGAACUGCGUCGAGCACACAAACGCAGCGCCA--------GAGGUCGCCACUCUACAUAAAUAGUUUGUGACCUUUCGCUUGUUUUACAAACAAGCU-CCAAACAGCUGGCGUCGA ..(((..((((((...........))))))(((((--------((((((((....(((......)))...)))))))...(((((((.....))))))).-........))))))))). ( -36.70) >consensus AACGGAACUGCGUGGAAC_CACAAACUCAGCGCCA________GAGGUCGCCACUCUACAUAAAUAGUUUGUGACCUUUCGCUUGUUUUACAAACAAGCU_CCAAACAGUUGGCGUCGC ........((((.......))))......((((((........((((((((....(((......)))...))))))))..(((((((.....)))))))...........))))))... (-23.37 = -24.73 + 1.36)


| Location | 12,642,183 – 12,642,293 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.42 |
| Mean single sequence MFE | -36.07 |
| Consensus MFE | -22.64 |
| Energy contribution | -23.50 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12642183 110 - 23771897 GCGACGCCAACUGUUCAC-UGCUUGUUUGUAAAACAAGCGAAAGGUCACAAACUAUUUAUGUAGAGUGGCGACCUC--------UGGCGCUCAGUUUGCGCGUUCCACGCAGUUCCGUU (.(((((.(((((..(.(-((((((((.....)))))))...(((((.((..((((....))))..))..))))).--------.)).)..))))).))(((.....))).))).)... ( -33.60) >DroVir_CAF1 6430 111 - 1 UCGACGCCAACUGUUUGG-GGCUUGUUUGUAAAACAACCAAAAGGUCACAAACUAUUUGUGUAGAGUGGCGACCUCU-UGCCUCUGGCGCAGUGUUUGU--GUU-CGCACA---CCAUU ...............(((-.(((((.(((.....))).)))..(..(((((((...((((((..((.(((((....)-)))).)).)))))).))))))--)..-)))...---))).. ( -31.10) >DroEre_CAF1 6505 110 - 1 GCGACGCCAACUGUUGGG-AGCUUGUUUGUAAAACAAGCGAAAGGUCACAAACUAUUUAUGUAGAGUGGCGACCUC--------UGGCGCUCAGUUUGCGAGUUCCACGCAGUUCCGUU ..((((..((((((..((-(((((((.....((((.((((..(((((.((..((((....))))..))..))))).--------...))))..)))))))))))))..)))))).)))) ( -38.30) >DroYak_CAF1 5915 110 - 1 GCGACGCCAGCUGUUGGG-AGCUUGUUUGUAAAACAAGCGAGAGGUCACAAACUAUUUAUGUAGAGUGGCGACCUC--------UGGCGCCCAGUUAGCGAGUGCCACGCAGUUCCGUU (((.(((..((((..(((-.(((((((((.....))))))(((((((.((..((((....))))..))..))))))--------)))).)))...))))..)))...)))......... ( -40.10) >DroMoj_CAF1 6989 106 - 1 -------AAACUGUUUGGGGGCUUGUUUGUAAAACAACCAAAAGGUCACAAACUAUUUGUGUAGAGUGGCGACCUCUUUGCCGCUGGCGCAGUGUUUGU--GUU-CGCACA---CCAUU -------........(((..(((((.(((.....))).)))..(..(((((((...((((((..((((((((.....)))))))).)))))).))))))--)..-)))...---))).. ( -35.10) >DroAna_CAF1 7049 110 - 1 UCGACGCCAGCUGUUUGG-AGCUUGUUUGUAAAACAAGCGAAAGGUCACAAACUAUUUAUGUAGAGUGGCGACCUC--------UGGCGCUGCGUUUGUGUGCUCGACGCAGUUCCGUU ..((((((((........-.(((((((.....)))))))...(((((.((..((((....))))..))..))))))--------))))((((((((.(....)..))))))))...))) ( -38.20) >consensus GCGACGCCAACUGUUUGG_AGCUUGUUUGUAAAACAAGCGAAAGGUCACAAACUAUUUAUGUAGAGUGGCGACCUC________UGGCGCUCAGUUUGCG_GUUCCACGCAGUUCCGUU ....(((((...........(((((((.....)))))))...(((((.((..((((....))))..))..))))).........)))))........(((.......)))......... (-22.64 = -23.50 + 0.86)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:08 2006