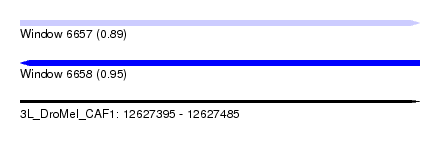
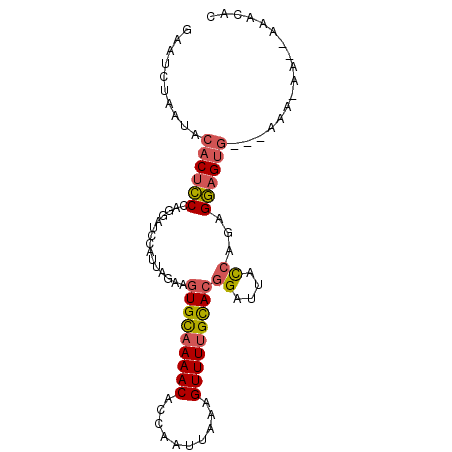
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,627,395 – 12,627,485 |
| Length | 90 |
| Max. P | 0.953781 |

| Location | 12,627,395 – 12,627,485 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 80.96 |
| Mean single sequence MFE | -22.66 |
| Consensus MFE | -14.76 |
| Energy contribution | -17.09 |
| Covariance contribution | 2.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.885117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12627395 90 + 23771897 GUGUUU--UU-UUUUUGCACUCCUCUGGUAAUCCGUGCAAAACUUUAAUUGGUGUUUUGCACUUCUAAUGGAUCCUGGGAGUGUAUUAGAUUC ......--.(-((..(((((((((..((..(((((((((((((..........))))))))).......)))))).)))))))))..)))... ( -29.41) >DroGri_CAF1 128234 80 + 1 GUUUUU--CU-UUU---GCCAACAAAAAUAAGCAAUACAAAACUUUAAUCGACGUUUUACAAUUCUAAUGU-----GGCAGUGCAUA--AUUC ......--..-.((---(((..................(((((..........)))))(((.......)))-----)))))......--.... ( -6.20) >DroSec_CAF1 115251 87 + 1 GCGUUU--UU-UUU---CACUCCUCUGGUAAUCCGUGCAAAACUUUAAUUGGUGUUUUGCACUUCUAAUGGAUCCUGGGAGUGUAUUAGAUUC ..((((--..-...---(((((((..((..(((((((((((((..........))))))))).......)))))).)))))))....)))).. ( -25.51) >DroSim_CAF1 118607 87 + 1 GCGUUU--UU-UUU---CACUCCUCUGGUAAUCCGUGCAAAACUUUAAUUGGUGUUUUGCACUUCUAAUGGAUCCUGGGAGUGUAUUAGAUUC ..((((--..-...---(((((((..((..(((((((((((((..........))))))))).......)))))).)))))))....)))).. ( -25.51) >DroEre_CAF1 119223 86 + 1 GUGUGUU----UUU---CACUCCUCUGGUAAUCCGUGCAAAACUUUAAUUGGUGUUUUGCACUUCUAAUGGAUCCUGGGAGUGUAUUAGAUUC ....(((----(..---(((((((..((..(((((((((((((..........))))))))).......)))))).)))))))....)))).. ( -25.31) >DroAna_CAF1 112208 90 + 1 GCUUUUUUGCUUGC---CACUCCGCUGGUAAUCCGUGCAAAACUUUAAUUGGUGUUUGGCACUUCUAAUGGAUCCUGGGAGUGUAUUAGGCCC ........(((((.---((((((.(.((..((((((((.((((..........)))).)))).......)))))).)))))))...))))).. ( -24.01) >consensus GCGUUU__UU_UUU___CACUCCUCUGGUAAUCCGUGCAAAACUUUAAUUGGUGUUUUGCACUUCUAAUGGAUCCUGGGAGUGUAUUAGAUUC .................((((((...((..(((((((((((((..........))))))))).......))))))..)))))).......... (-14.76 = -17.09 + 2.33)

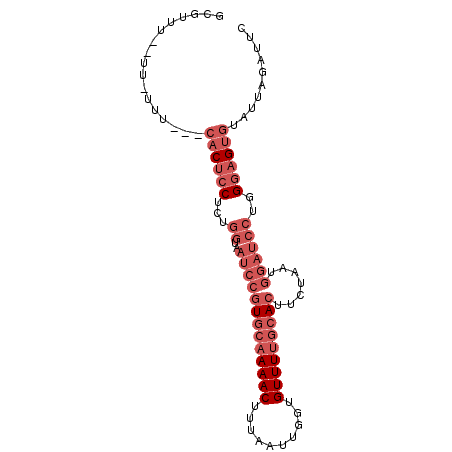
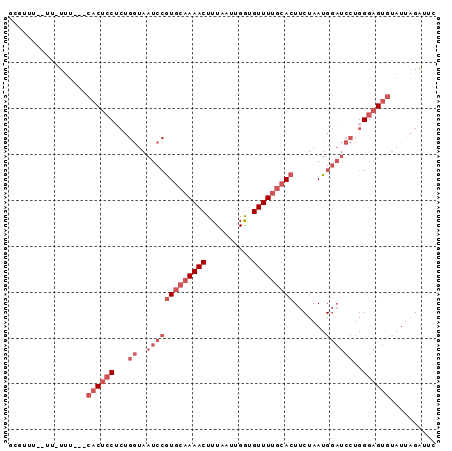
| Location | 12,627,395 – 12,627,485 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 80.96 |
| Mean single sequence MFE | -21.19 |
| Consensus MFE | -15.94 |
| Energy contribution | -16.25 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.953781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12627395 90 - 23771897 GAAUCUAAUACACUCCCAGGAUCCAUUAGAAGUGCAAAACACCAAUUAAAGUUUUGCACGGAUUACCAGAGGAGUGCAAAAA-AA--AAACAC ..........(((((((.((((((.......(((((((((..........)))))))))))))..)).).))))))......-..--...... ( -23.31) >DroGri_CAF1 128234 80 - 1 GAAU--UAUGCACUGCC-----ACAUUAGAAUUGUAAAACGUCGAUUAAAGUUUUGUAUUGCUUAUUUUUGUUGGC---AAA-AG--AAAAAC ....--.......((((-----(.((.(((((.((((.(((..(((....))).))).))))..))))).))))))---)..-..--...... ( -10.80) >DroSec_CAF1 115251 87 - 1 GAAUCUAAUACACUCCCAGGAUCCAUUAGAAGUGCAAAACACCAAUUAAAGUUUUGCACGGAUUACCAGAGGAGUG---AAA-AA--AAACGC ..........(((((((.((((((.......(((((((((..........)))))))))))))..)).).))))))---...-..--...... ( -24.01) >DroSim_CAF1 118607 87 - 1 GAAUCUAAUACACUCCCAGGAUCCAUUAGAAGUGCAAAACACCAAUUAAAGUUUUGCACGGAUUACCAGAGGAGUG---AAA-AA--AAACGC ..........(((((((.((((((.......(((((((((..........)))))))))))))..)).).))))))---...-..--...... ( -24.01) >DroEre_CAF1 119223 86 - 1 GAAUCUAAUACACUCCCAGGAUCCAUUAGAAGUGCAAAACACCAAUUAAAGUUUUGCACGGAUUACCAGAGGAGUG---AAA----AACACAC ..........(((((((.((((((.......(((((((((..........)))))))))))))..)).).))))))---...----....... ( -24.01) >DroAna_CAF1 112208 90 - 1 GGGCCUAAUACACUCCCAGGAUCCAUUAGAAGUGCCAAACACCAAUUAAAGUUUUGCACGGAUUACCAGCGGAGUG---GCAAGCAAAAAAGC ..((......(((((((.((((((.......((((.((((..........)))).))))))))..)).).))))))---....))........ ( -21.01) >consensus GAAUCUAAUACACUCCCAGGAUCCAUUAGAAGUGCAAAACACCAAUUAAAGUUUUGCACGGAUUACCAGAGGAGUG___AAA_AA__AAACAC ..........((((((...............(((((((((..........)))))))))((....))...))))))................. (-15.94 = -16.25 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:04 2006