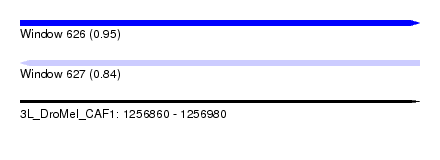
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,256,860 – 1,256,980 |
| Length | 120 |
| Max. P | 0.945502 |

| Location | 1,256,860 – 1,256,980 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.25 |
| Mean single sequence MFE | -37.90 |
| Consensus MFE | -29.00 |
| Energy contribution | -29.33 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
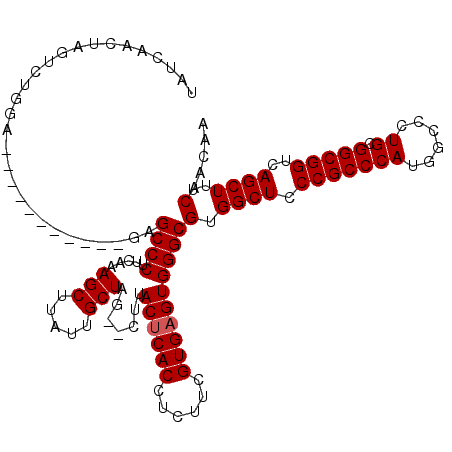
>3L_DroMel_CAF1 1256860 120 + 23771897 UAUCAACUAGUCUGGAGGUGGCAGGUAGAGCCCUUUAAAGCUUAUUGCUAGUGUUUACCCACCACUUCGUGAGUGGGCGUGGCUGCCGCCCAUGGCCCUGCGGCGGUCAGCUUUCAACAA ......(((.((..(((((((..((((((.(.((....(((.....))))).)))))))..)))))))..)).)))((.((((((((((..........))))))))))))......... ( -43.50) >DroSec_CAF1 11648 107 + 1 UAUCAACUAGUCUGGA-----------GUGCCCUUCAAAGCUUAUUGCUAG--CUUACUCACCUCUUCGUGAGUGGGCGUGGCUCCCGCCCAUGGCCCUGCGGCGGUCAGCUUUCAACAA .........((..(((-----------((((((.....(((.....))).(--(((((((((......))))))))))).)))..(((((((......)).)))))...)))))..)).. ( -36.30) >DroSim_CAF1 11610 91 + 1 ---------------------------CAGCCCUUCAAAGCUUAUUGCUAG--CUUACUCACCUCUUCGUGAGUGGGCGUGGCUCCCGCCCAUGGCCCUGCGGCGGUCAGCUUUCAACAA ---------------------------.........((((((....(((((--(((((((((......)))))))))).))))..(((((((......)).)))))..))))))...... ( -33.90) >consensus UAUCAACUAGUCUGGA___________GAGCCCUUCAAAGCUUAUUGCUAG__CUUACUCACCUCUUCGUGAGUGGGCGUGGCUCCCGCCCAUGGCCCUGCGGCGGUCAGCUUUCAACAA .............................((((.....(((.....))).......((((((......))))))))))(.((((.(((((((......)).)))))..))))..)..... (-29.00 = -29.33 + 0.33)

| Location | 1,256,860 – 1,256,980 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.25 |
| Mean single sequence MFE | -36.77 |
| Consensus MFE | -31.32 |
| Energy contribution | -31.10 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
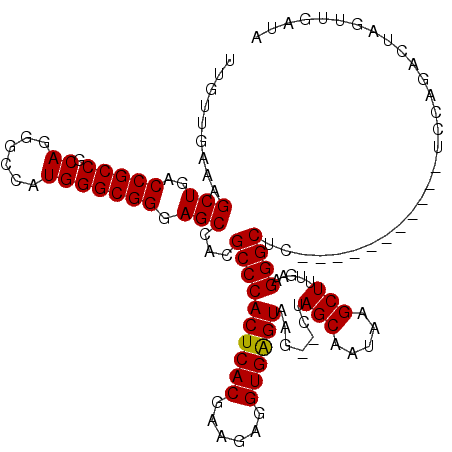
>3L_DroMel_CAF1 1256860 120 - 23771897 UUGUUGAAAGCUGACCGCCGCAGGGCCAUGGGCGGCAGCCACGCCCACUCACGAAGUGGUGGGUAAACACUAGCAAUAAGCUUUAAAGGGCUCUACCUGCCACCUCCAGACUAGUUGAUA .((((....((((.(((((.((......)))))))))))...((((((.(((...))))))))).))))....((((.((((......(((.......)))......)).)).))))... ( -39.50) >DroSec_CAF1 11648 107 - 1 UUGUUGAAAGCUGACCGCCGCAGGGCCAUGGGCGGGAGCCACGCCCACUCACGAAGAGGUGAGUAAG--CUAGCAAUAAGCUUUGAAGGGCAC-----------UCCAGACUAGUUGAUA .(((..(.(((((.(((((.((......))))))).......((((((((((......))))))(((--((.......)))))....))))..-----------..))).))..)..))) ( -36.00) >DroSim_CAF1 11610 91 - 1 UUGUUGAAAGCUGACCGCCGCAGGGCCAUGGGCGGGAGCCACGCCCACUCACGAAGAGGUGAGUAAG--CUAGCAAUAAGCUUUGAAGGGCUG--------------------------- .(((((..((((..........((((....(((....)))..))))((((((......)))))).))--))..)))))(((((....))))).--------------------------- ( -34.80) >consensus UUGUUGAAAGCUGACCGCCGCAGGGCCAUGGGCGGGAGCCACGCCCACUCACGAAGAGGUGAGUAAG__CUAGCAAUAAGCUUUGAAGGGCUC___________UCCAGACUAGUUGAUA .........(((..(((((.((......))))))).)))...((((((((((......)))))).......(((.....))).....))))............................. (-31.32 = -31.10 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:37 2006