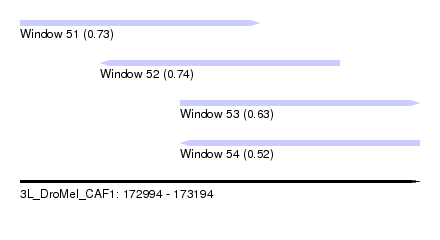
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 172,994 – 173,194 |
| Length | 200 |
| Max. P | 0.744504 |

| Location | 172,994 – 173,114 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -39.03 |
| Consensus MFE | -36.27 |
| Energy contribution | -36.83 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 172994 120 + 23771897 UAAGAGUGCUAAAAUUACUUGUCAAACGCUUUUUAGGUGGACAAGCCCUUCGGGUCAUUUCAUAACUGUAGUUUUGGCCCUCGUCAACGCUUGAAACACCUGAAGGUGUCUCGUUGGCCG ...(((.((((((((((((((((...((((.....)))))))))(.((....)).)...........))))))))))).)))(((((((......(((((....)))))..))))))).. ( -40.60) >DroSec_CAF1 6700 120 + 1 UAAGAGUGCUAAAAUUGCUGGUCAAACGCUUUUUAGGUGGACAAGCCCUUCGGGUCAUCUCAUAACUGUAGUUUUGGCCCUCGUCGACGCUUGAAACACCUGAUGGUGUCUCGUUGGCCG ...(((.(((((((((((.((((...((((.....)))))))..(.((....)).).........).))))))))))).)))(((((((......(((((....)))))..))))))).. ( -39.10) >DroYak_CAF1 6909 120 + 1 UAAGAGUGCUAUAAUUGCUUGUCAAACGCUUCUUUGGUGGACAAGCCCUUCGGGUCAUCUCAUAACUGUAGUUUUGGCCCUCGUCGACGCUUGAAACCCCUGAAGGUGUCUCGUUGGCCG .......((((((...(((((((...((((.....)))))))))))((....))............))))))...((((..((..(((((((...........))))))).))..)))). ( -37.40) >consensus UAAGAGUGCUAAAAUUGCUUGUCAAACGCUUUUUAGGUGGACAAGCCCUUCGGGUCAUCUCAUAACUGUAGUUUUGGCCCUCGUCGACGCUUGAAACACCUGAAGGUGUCUCGUUGGCCG ...(((.((((((((((((((((...((((.....)))))))))((((...))))............))))))))))).)))(((((((......(((((....)))))..))))))).. (-36.27 = -36.83 + 0.56)

| Location | 173,034 – 173,154 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -41.00 |
| Consensus MFE | -36.93 |
| Energy contribution | -37.93 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 173034 120 - 23771897 CACCAGGCCCCUAAGCUGAAGAUCAAAAUCAGGGGUUUGACGGCCAACGAGACACCUUCAGGUGUUUCAAGCGUUGACGAGGGCCAAAACUACAGUUAUGAAAUGACCCGAAGGGCUUGU ...(((((((.....((((.........))))(((((....((((...((((((((....))))))))...((....))..))))..(((....))).......)))))...))))))). ( -42.10) >DroSec_CAF1 6740 120 - 1 CACCAGGCCCCCAAGCUGAAGAUCAAAAUCAGGGGGUUGACGGCCAACGAGACACCAUCAGGUGUUUCAAGCGUCGACGAGGGCCAAAACUACAGUUAUGAGAUGACCCGAAGGGCUUGU ...(((((((.....((((.........)))).(((((...((((...((((((((....))))))))...((....))..))))....((.((....))))..)))))...))))))). ( -42.70) >DroYak_CAF1 6949 120 - 1 CACCAGGCCCCCAAGCUGAAGAUCAAAAUAAGGGGUUUGGCGGCCAACGAGACACCUUCAGGGGUUUCAAGCGUCGACGAGGGCCAAAACUACAGUUAUGAGAUGACCCGAAGGGCUUGU .....((((.(((((((................))))))).)))).(((((...(((((.(((((((((.(.(((......))))..(((....))).))))))..)))))))).))))) ( -38.19) >consensus CACCAGGCCCCCAAGCUGAAGAUCAAAAUCAGGGGUUUGACGGCCAACGAGACACCUUCAGGUGUUUCAAGCGUCGACGAGGGCCAAAACUACAGUUAUGAGAUGACCCGAAGGGCUUGU ...(((((((.....((((.........))))(((((....((((...((((((((....))))))))...((....))..))))..(((....))).......)))))...))))))). (-36.93 = -37.93 + 1.00)

| Location | 173,074 – 173,194 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -41.15 |
| Consensus MFE | -38.29 |
| Energy contribution | -38.63 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 173074 120 + 23771897 UCGUCAACGCUUGAAACACCUGAAGGUGUCUCGUUGGCCGUCAAACCCCUGAUUUUGAUCUUCAGCUUAGGGGCCUGGUGGUCAUCCUCGCUGCUUUCGAGAGUCUUACUGCGUCCUCGC .((...((((..((.(((((....))))).))..(((((..((..(((((((..(((.....))).)))))))..))..)))))..((((.......)))).........))))...)). ( -38.60) >DroSec_CAF1 6780 120 + 1 UCGUCGACGCUUGAAACACCUGAUGGUGUCUCGUUGGCCGUCAACCCCCUGAUUUUGAUCUUCAGCUUGGGGGCCUGGUGGUCAUCCUCACUGCUUUCGAGAGUCUUACUGCGUCCUCGC .((..(((((......((((....))))(((((.(((((..((..((((..(..(((.....))).)..))))..))..))))).............)))))........)))))..)). ( -40.84) >DroYak_CAF1 6989 120 + 1 UCGUCGACGCUUGAAACCCCUGAAGGUGUCUCGUUGGCCGCCAAACCCCUUAUUUUGAUCUUCAGCUUGGGGGCCUGGUGGUCAUCCUCACUGCUUUCGAGAGUCUUACUGCGUCCUCGC .((..(((((.(((.((.(.(((((((((.....(((((((((..(((((....(((.....)))...)))))..))))))))).....)).))))))).).)).)))..)))))..)). ( -44.00) >consensus UCGUCGACGCUUGAAACACCUGAAGGUGUCUCGUUGGCCGUCAAACCCCUGAUUUUGAUCUUCAGCUUGGGGGCCUGGUGGUCAUCCUCACUGCUUUCGAGAGUCUUACUGCGUCCUCGC .((..(((((.(((.((.(.(((((((((.....(((((((((..(((((((..(((.....))).)))))))..))))))))).....)).))))))).).)).)))..)))))..)). (-38.29 = -38.63 + 0.34)


| Location | 173,074 – 173,194 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -40.70 |
| Consensus MFE | -36.74 |
| Energy contribution | -36.97 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.90 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 173074 120 - 23771897 GCGAGGACGCAGUAAGACUCUCGAAAGCAGCGAGGAUGACCACCAGGCCCCUAAGCUGAAGAUCAAAAUCAGGGGUUUGACGGCCAACGAGACACCUUCAGGUGUUUCAAGCGUUGACGA .((..(((((.((.....(((((.......))))).((.((..(((((((((.(..((.....))...).)))))))))..)).))))((((((((....))))))))..)))))..)). ( -41.70) >DroSec_CAF1 6780 120 - 1 GCGAGGACGCAGUAAGACUCUCGAAAGCAGUGAGGAUGACCACCAGGCCCCCAAGCUGAAGAUCAAAAUCAGGGGGUUGACGGCCAACGAGACACCAUCAGGUGUUUCAAGCGUCGACGA .((..(((((.((..(((((((.....((((..((..(.((....)))..))..))))..(((....))).)))))))....))....((((((((....))))))))..)))))..)). ( -39.60) >DroYak_CAF1 6989 120 - 1 GCGAGGACGCAGUAAGACUCUCGAAAGCAGUGAGGAUGACCACCAGGCCCCCAAGCUGAAGAUCAAAAUAAGGGGUUUGGCGGCCAACGAGACACCUUCAGGGGUUUCAAGCGUCGACGA .((..(((((.((....(((.(....)....)))..((.((.(((((((((.....((.....))......))))))))).)).))))(((((.((.....)))))))..)))))..)). ( -40.80) >consensus GCGAGGACGCAGUAAGACUCUCGAAAGCAGUGAGGAUGACCACCAGGCCCCCAAGCUGAAGAUCAAAAUCAGGGGUUUGACGGCCAACGAGACACCUUCAGGUGUUUCAAGCGUCGACGA .((..(((((.((.....(((((.......))))).((.((..((((((((.....((.....))......))))))))..)).))))((((((((....))))))))..)))))..)). (-36.74 = -36.97 + 0.23)

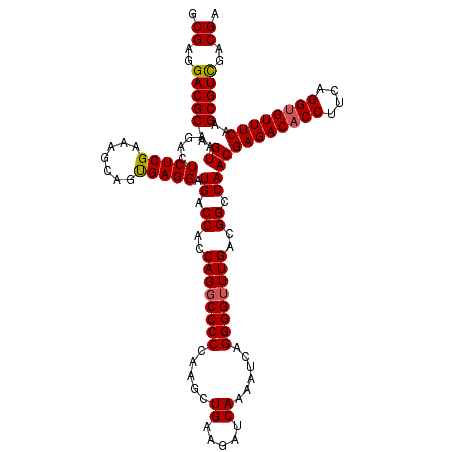
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:43 2006