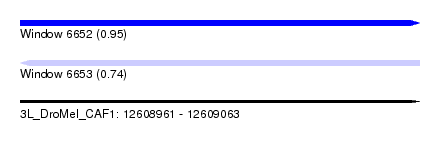
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,608,961 – 12,609,063 |
| Length | 102 |
| Max. P | 0.952578 |

| Location | 12,608,961 – 12,609,063 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 89.84 |
| Mean single sequence MFE | -25.37 |
| Consensus MFE | -20.90 |
| Energy contribution | -21.98 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
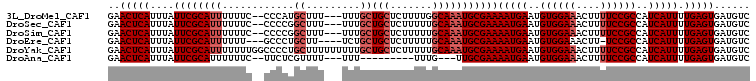
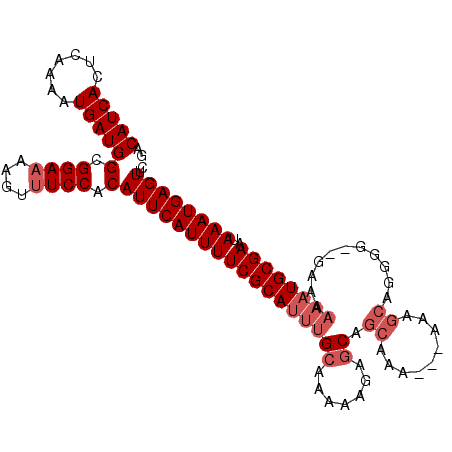
>3L_DroMel_CAF1 12608961 102 + 23771897 GAACUCAUUUAUUCGCAUUUUUUC--CCCAUGCUUU---UUUGCUGCUCUUUUGGCAAAUGCGAAAAUGAAUGUGGAAACUUUUCCGCCAUCAUUUUGAGUGAUGUC ..(((((....(((((((((....--.....((...---...)).(((.....))))))))))))(((((..((((((....))))))..))))).)))))...... ( -26.70) >DroSec_CAF1 96827 102 + 1 GAACUCAUUUAUUCGCAUUUUUUC--CCCCGGCUUU---UUUGCUGCUCUUUUUGCAAAUGCGAAAAUGAAUGUGGAAACUUUUCCGCCAUCAUUUUGAGUGAUGUC ..(((((....(((((((((....--....(((...---...)))((.......)))))))))))(((((..((((((....))))))..))))).)))))...... ( -27.70) >DroSim_CAF1 99712 102 + 1 GAACUCAUUUAUUCGCAUUUUUUC--CCCCGGCUUU---UUUGCUGCUCUUUUUGCAAAUGCGAAAAUGAAUGUGGAAACUUUUCCGCCAUCAUUUUGAGUGAUGUC ..(((((....(((((((((....--....(((...---...)))((.......)))))))))))(((((..((((((....))))))..))))).)))))...... ( -27.70) >DroEre_CAF1 99981 99 + 1 GAACUCAUUUAUUCGCAUUUUUU---GCCCUGCUU----UCUGCUGCUCUUUUUGCAAAUGCGAAAAUGAAUGUGGAAACUU-UCCGCCAUCAUUUUGAGUGAUGUC ..(((((....(((((((((...---.....((..----...)).((.......)))))))))))(((((..(((((.....-)))))..))))).)))))...... ( -25.20) >DroYak_CAF1 100755 107 + 1 GAACUCAUUUAUUCGCAUUUUUUUGGCCCCUGCUUUUUUUUUGCUGCUCUUUUUGCAAAUGCGAAAAUGAAUGUGGAAACUUUUCCGCCAUCAUUUUGAGUGAUGUC ..(((((....(((((((((....(((....)))...........((.......)))))))))))(((((..((((((....))))))..))))).)))))...... ( -25.60) >DroAna_CAF1 93836 90 + 1 GAACUCAUUUAUUCGCAUUUUUUC--UUCUCGUUUU---UUU---------UUUG---UUGCGAAAAUGAAUGUGGAAACUUUUCCGCCAUCAUUUUGAGUGAUGUC ..(((((....((((((.......--..........---...---------....---.))))))(((((..((((((....))))))..))))).)))))...... ( -19.31) >consensus GAACUCAUUUAUUCGCAUUUUUUC__CCCCUGCUUU___UUUGCUGCUCUUUUUGCAAAUGCGAAAAUGAAUGUGGAAACUUUUCCGCCAUCAUUUUGAGUGAUGUC ..(((((....((((((((............((.........))(((.......)))))))))))(((((..((((((....))))))..))))).)))))...... (-20.90 = -21.98 + 1.08)
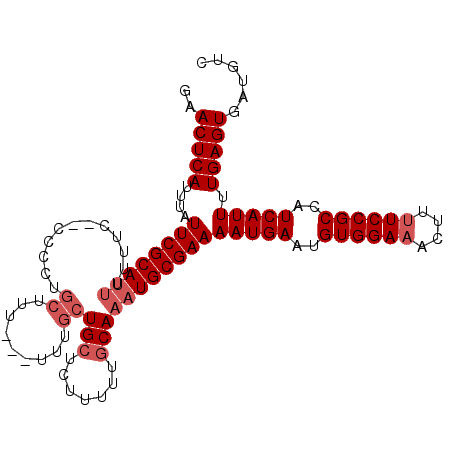
| Location | 12,608,961 – 12,609,063 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 89.84 |
| Mean single sequence MFE | -21.83 |
| Consensus MFE | -17.96 |
| Energy contribution | -19.17 |
| Covariance contribution | 1.21 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12608961 102 - 23771897 GACAUCACUCAAAAUGAUGGCGGAAAAGUUUCCACAUUCAUUUUCGCAUUUGCCAAAAGAGCAGCAAA---AAAGCAUGGG--GAAAAAAUGCGAAUAAAUGAGUUC ..(((((.......)))))(.((((....)))).)((((((((((((((((.((......((......---...))....)--)...))))))))..)))))))).. ( -24.00) >DroSec_CAF1 96827 102 - 1 GACAUCACUCAAAAUGAUGGCGGAAAAGUUUCCACAUUCAUUUUCGCAUUUGCAAAAAGAGCAGCAAA---AAAGCCGGGG--GAAAAAAUGCGAAUAAAUGAGUUC ..(((((.......)))))(.((((....)))).)((((((((((((((((((.......)).((...---...)).....--....))))))))..)))))))).. ( -22.90) >DroSim_CAF1 99712 102 - 1 GACAUCACUCAAAAUGAUGGCGGAAAAGUUUCCACAUUCAUUUUCGCAUUUGCAAAAAGAGCAGCAAA---AAAGCCGGGG--GAAAAAAUGCGAAUAAAUGAGUUC ..(((((.......)))))(.((((....)))).)((((((((((((((((((.......)).((...---...)).....--....))))))))..)))))))).. ( -22.90) >DroEre_CAF1 99981 99 - 1 GACAUCACUCAAAAUGAUGGCGGA-AAGUUUCCACAUUCAUUUUCGCAUUUGCAAAAAGAGCAGCAGA----AAGCAGGGC---AAAAAAUGCGAAUAAAUGAGUUC ..(((((.......)))))(.(((-.....))).)((((((((((((((((((.......)).((...----..)).....---...))))))))..)))))))).. ( -22.70) >DroYak_CAF1 100755 107 - 1 GACAUCACUCAAAAUGAUGGCGGAAAAGUUUCCACAUUCAUUUUCGCAUUUGCAAAAAGAGCAGCAAAAAAAAAGCAGGGGCCAAAAAAAUGCGAAUAAAUGAGUUC ..(((((.......)))))(.((((....)))).)((((((((((((((((((.......)).((.........))...........))))))))..)))))))).. ( -22.80) >DroAna_CAF1 93836 90 - 1 GACAUCACUCAAAAUGAUGGCGGAAAAGUUUCCACAUUCAUUUUCGCAA---CAAA---------AAA---AAAACGAGAA--GAAAAAAUGCGAAUAAAUGAGUUC ..(((((.......)))))(.((((....)))).)(((((((((((((.---....---------...---..........--.......)))))..)))))))).. ( -15.71) >consensus GACAUCACUCAAAAUGAUGGCGGAAAAGUUUCCACAUUCAUUUUCGCAUUUGCAAAAAGAGCAGCAAA___AAAGCAGGGG__GAAAAAAUGCGAAUAAAUGAGUUC ..(((((.......)))))(.((((....)))).)((((((((((((((((((.......)).((.........))...........))))))))..)))))))).. (-17.96 = -19.17 + 1.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:59 2006