
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,608,316 – 12,608,453 |
| Length | 137 |
| Max. P | 0.997991 |

| Location | 12,608,316 – 12,608,424 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.94 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -20.88 |
| Energy contribution | -21.12 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12608316 108 + 23771897 AGAUGCUAUUCAAUUCAAAUGGCGUGGGUUGCAUGUGAAAUGGCACGCACAUGCGAAGGUGUGCCCUAUACCUAAAUAUAAUACC-CCUUAUUCCAUAUCCACAUC------CAG--- .(((((((((.......))))))((((((((((((((..........)))))))))(((((((...)))))))............-............))))))))------...--- ( -28.10) >DroSec_CAF1 96193 98 + 1 AGAUGCUAUUCAAUUCAAAUGGCGUGGGUUGCAUGUGAAGUGGCACGCACAUGCUAAGGUGCGCCCCAUACCU----GUAAUACC-CCUUAUGCCAUAU------C------CAG--- ..................(((((((((((((((.((...((((..(((((........)))))..)))))).)----)))..)))-)...)))))))..------.------...--- ( -28.70) >DroSim_CAF1 99073 104 + 1 AGAUGCUAUUCAAUUCAAAUGGCGUGGGUUGCAUGUGAAAUGGCACGCACAUGCGAAGGUGCGCCCCAUACCU----GUAAUACC-CCUUAUUCCAUAUCCACAUC------CAG--- .(((((((((.......))))))((((((((((((((..........)))))))))(((((.......)))))----........-............))))))))------...--- ( -26.10) >DroEre_CAF1 99394 104 + 1 AGAUGCCAUUCAAUUCAAAUGGCGUGGGAUGCACGUGGAAUGGCACGCACAUGCGAAGGUGUGCCGCACACCC----G---UACU-CCUUAUGCCACAUCCACAUC------CUGAUC ..((((((((.......))))))))(((((....(((((.((((((((....)))((((.((((.(.....).----)---))).-)))).)))))..))))))))------)).... ( -37.90) >DroYak_CAF1 100124 108 + 1 AGAUGCUAUUCAAUUCAAAUGGCGUGGGAUGCACGUGAAGUGGCACGCACAUGCUAAGGUGUACC---CAUCC----A---UACCCCCCUAUGCCACAUCCACAUCUAGAUUCAGAUC ((((((((((.......))))))(((((((....))...((((((.((....))...(((((...---.....----)---))))......)))))).)))))))))........... ( -26.30) >consensus AGAUGCUAUUCAAUUCAAAUGGCGUGGGUUGCAUGUGAAAUGGCACGCACAUGCGAAGGUGUGCCCCAUACCU____GUAAUACC_CCUUAUGCCAUAUCCACAUC______CAG___ ...................(((((((((..(((((((..........)))))))...((((.......))))...............)))))))))...................... (-20.88 = -21.12 + 0.24)



| Location | 12,608,354 – 12,608,453 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.61 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -16.58 |
| Energy contribution | -16.70 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.997991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12608354 99 + 23771897 AAUGGCACGCACAUGCGAAGGUGUGCCCUAUACCUAAAUAUAAUACC-CCUUAUUCCAUAUCCACAUC------CAG------AUCCCAAUCCAUAUCUAUGUGGAUACGCU ...(((.(((....))).(((((((...)))))))............-..........(((((((((.------.((------((..........))))))))))))).))) ( -22.30) >DroSec_CAF1 96231 89 + 1 AGUGGCACGCACAUGCUAAGGUGCGCCCCAUACCU----GUAAUACC-CCUUAUGCCAUAU------C------CAG------AUCCAAAUCCAUAUCUAUGUGGAUACGGU .((((..(((((........)))))..))))....----........-......(((.(((------(------((.------((..............)).)))))).))) ( -21.44) >DroSim_CAF1 99111 95 + 1 AAUGGCACGCACAUGCGAAGGUGCGCCCCAUACCU----GUAAUACC-CCUUAUUCCAUAUCCACAUC------CAG------AUCCAAAUCCAUAUCUAUGUGGAUACGGU .((((..(((((........)))))..))))....----........-.......((.(((((((((.------.((------((..........))))))))))))).)). ( -25.70) >DroEre_CAF1 99432 98 + 1 AAUGGCACGCACAUGCGAAGGUGUGCCGCACACCC----G---UACU-CCUUAUGCCACAUCCACAUC------CUGAUCCGAAUCCGAAUCCAUAUCUAUGUGGAUACGGU ..((((((((....)))((((.((((.(.....).----)---))).-)))).)))))..........------...........(((.(((((((....))))))).))). ( -29.40) >DroYak_CAF1 100162 102 + 1 AGUGGCACGCACAUGCUAAGGUGUACC---CAUCC----A---UACCCCCCUAUGCCACAUCCACAUCUAGAUUCAGAUCCAAAUCCGAAUCCAUAUCUAUGUGGAUACGGU .((((((.((....))...(((((...---.....----)---))))......))))))((((((((...(((((.(((....))).))))).......))))))))..... ( -29.50) >consensus AAUGGCACGCACAUGCGAAGGUGUGCCCCAUACCU____GUAAUACC_CCUUAUGCCAUAUCCACAUC______CAG______AUCCAAAUCCAUAUCUAUGUGGAUACGGU .((((((.((....))...((((.......))))...................))))))..........................((..(((((((....)))))))..)). (-16.58 = -16.70 + 0.12)

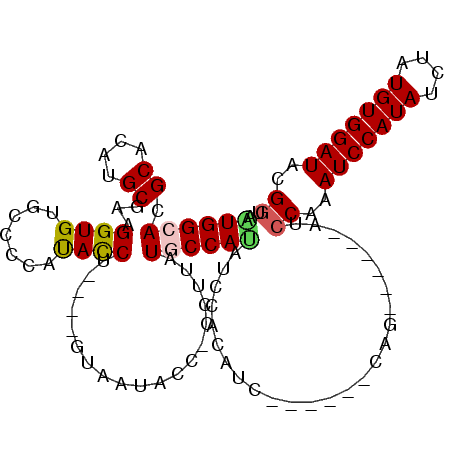

| Location | 12,608,354 – 12,608,453 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.61 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -18.13 |
| Energy contribution | -18.89 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12608354 99 - 23771897 AGCGUAUCCACAUAGAUAUGGAUUGGGAU------CUG------GAUGUGGAUAUGGAAUAAGG-GGUAUUAUAUUUAGGUAUAGGGCACACCUUCGCAUGUGCGUGCCAUU ..(((((((((((.....(((((....))------)))------.)))))))))))........-((((((((((....)))))..(((((........))))))))))... ( -28.70) >DroSec_CAF1 96231 89 - 1 ACCGUAUCCACAUAGAUAUGGAUUUGGAU------CUG------G------AUAUGGCAUAAGG-GGUAUUAC----AGGUAUGGGGCGCACCUUAGCAUGUGCGUGCCACU .(((((((......)))))))......((------(((------.------..(((.(......-).)))..)----)))).(((.((((((........)))))).))).. ( -28.20) >DroSim_CAF1 99111 95 - 1 ACCGUAUCCACAUAGAUAUGGAUUUGGAU------CUG------GAUGUGGAUAUGGAAUAAGG-GGUAUUAC----AGGUAUGGGGCGCACCUUCGCAUGUGCGUGCCAUU .((((((((((((.....(((((....))------)))------.)))))))))))).......-........----....((((.((((((........)))))).)))). ( -35.40) >DroEre_CAF1 99432 98 - 1 ACCGUAUCCACAUAGAUAUGGAUUCGGAUUCGGAUCAG------GAUGUGGAUGUGGCAUAAGG-AGUA---C----GGGUGUGCGGCACACCUUCGCAUGUGCGUGCCAUU .((((((((((((.......((((((....))))))..------.))))))))))))......(-.(((---(----(.(..(((((.......)))))..).))))))... ( -37.00) >DroYak_CAF1 100162 102 - 1 ACCGUAUCCACAUAGAUAUGGAUUCGGAUUUGGAUCUGAAUCUAGAUGUGGAUGUGGCAUAGGGGGGUA---U----GGAUG---GGUACACCUUAGCAUGUGCGUGCCACU .....((((((((.....(((((((((((....))))))))))).))))))))(((((((..(((((((---(----.....---.)))).)))).((....))))))))). ( -42.20) >consensus ACCGUAUCCACAUAGAUAUGGAUUCGGAU______CUG______GAUGUGGAUAUGGCAUAAGG_GGUAUUAC____AGGUAUGGGGCACACCUUCGCAUGUGCGUGCCAUU .((((((((((((................................))))))))))))............................(((((.((.......).).)))))... (-18.13 = -18.89 + 0.76)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:56 2006