
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,604,108 – 12,604,243 |
| Length | 135 |
| Max. P | 0.898378 |

| Location | 12,604,108 – 12,604,203 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 86.48 |
| Mean single sequence MFE | -17.44 |
| Consensus MFE | -14.21 |
| Energy contribution | -14.53 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12604108 95 + 23771897 UAAAGAAUUU-UUUCAUAAAUCUCCGCAUUUGCAUUAUUUUCGACUCAAGCAAUAUAUAGUAAUUAGAGUCGAAAAAGGCGAACAUGCAACAAUUA ....(((...-.)))..........((((((((....((((((((((...................))))))))))..))))..))))........ ( -19.91) >DroSec_CAF1 91984 93 + 1 UAAAGAAUUUUUUUCAUGAAUCUCCGCAUUUGCAUUAUUUUUGACUCGAGAAGUAUA---UAAUUAGAGUCGAAAAAGGCGAACAUGCAACAAUUA ....(((.....)))..........((((((((....((((((((((...((.....---...)).))))))))))..))))..))))........ ( -18.70) >DroSim_CAF1 94793 91 + 1 UAAAGAAUUUUUUUCAUGAAUCUCCGCAUUUGCAUUAUUUUCGACUCGAGAA--AUA---UAAUUAGAGUCGAAAAAGGCGAACAUGCAACAAUUA ....(((.....)))..........((((((((....((((((((((.....--...---......))))))))))..))))..))))........ ( -20.94) >DroEre_CAF1 95247 87 + 1 UGAAGAAUUC--UUCAUGAAUCUCCGUAUUUGCAUUAUUUUCGACUUAAGCA--AUA---UAAUUAAAGCCAAA--AGGCGAACAUGCAACUUCUA .((((.((((--.....))))........((((((....((((.(((..((.--...---........))...)--)).)))).)))))))))).. ( -12.90) >DroYak_CAF1 95644 89 + 1 UAAAGAAUUU--UUCAUGAAUCUCCGCAUUUGCAUUAUUUUCGACUUAAGCA--AUA---UAAUUAAAGUCAAAAUAGGCAAGCAUGCAACAUCUA ..........--.............((((((((.(((((((.(((((.....--...---......))))))))))))))))..))))........ ( -14.74) >consensus UAAAGAAUUU_UUUCAUGAAUCUCCGCAUUUGCAUUAUUUUCGACUCAAGCA__AUA___UAAUUAGAGUCGAAAAAGGCGAACAUGCAACAAUUA ....(((.....)))..........((((((((....((((((((((...................))))))))))..))))..))))........ (-14.21 = -14.53 + 0.32)



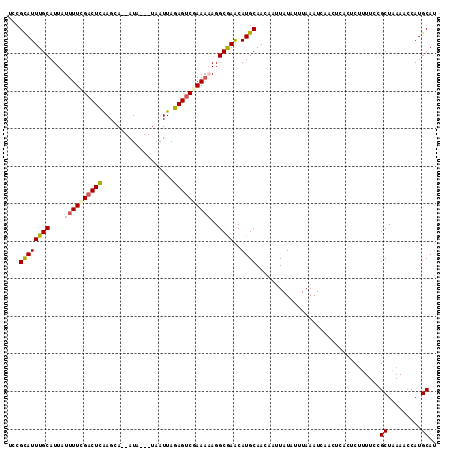
| Location | 12,604,129 – 12,604,243 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 86.65 |
| Mean single sequence MFE | -17.70 |
| Consensus MFE | -14.95 |
| Energy contribution | -15.27 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12604129 114 + 23771897 UCCGCAUUUGCAUUAUUUUCGACUCAAGCAAUAUAUAGUAAUUAGAGUCGAAAAAGGCGAACAUGCAACAAUUAUAUUUAAAUCAUCUCACUCUUUUCCGCUAAAACCAUGCAU ...((((((((....((((((((((...................))))))))))..))))..)))).................................((.........)).. ( -20.21) >DroSec_CAF1 92006 111 + 1 UCCGCAUUUGCAUUAUUUUUGACUCGAGAAGUAUA---UAAUUAGAGUCGAAAAAGGCGAACAUGCAACAAUUAUAUUUAAAUCAUCGCACUCUUUUCCGCUAAAACCAUGCAU ...((((((((....((((((((((...((.....---...)).))))))))))..))))..)))).....................(((...((((.....))))...))).. ( -19.20) >DroSim_CAF1 94815 109 + 1 UCCGCAUUUGCAUUAUUUUCGACUCGAGAA--AUA---UAAUUAGAGUCGAAAAAGGCGAACAUGCAACAAUUAUAUUUAAAUCUACUCACUCUUUUCCGCUAAAACCAUGCAU ...((((((((....((((((((((.....--...---......))))))))))..))))..)))).................................((.........)).. ( -21.24) >DroEre_CAF1 95267 107 + 1 UCCGUAUUUGCAUUAUUUUCGACUUAAGCA--AUA---UAAUUAAAGCCAAA--AGGCGAACAUGCAACUUCUAUAUUUAAAUCGCCUCACUCAAUUCCGCUUAAACCAUGCAU ........(((((.......(..((((((.--...---........(((...--.)))(((........)))...........................))))))..)))))). ( -12.11) >DroYak_CAF1 95664 109 + 1 UCCGCAUUUGCAUUAUUUUCGACUUAAGCA--AUA---UAAUUAAAGUCAAAAUAGGCAAGCAUGCAACAUCUAUAUUUAAAUAAACUCACUCUAUUCCGCUUAAGCCAUGCAA ...((((((((.(((((((.(((((.....--...---......))))))))))))))))..)))).................................((.........)).. ( -15.74) >consensus UCCGCAUUUGCAUUAUUUUCGACUCAAGCA__AUA___UAAUUAGAGUCGAAAAAGGCGAACAUGCAACAAUUAUAUUUAAAUCAACUCACUCUUUUCCGCUAAAACCAUGCAU ...((((((((....((((((((((...................))))))))))..))))..)))).................................((.........)).. (-14.95 = -15.27 + 0.32)


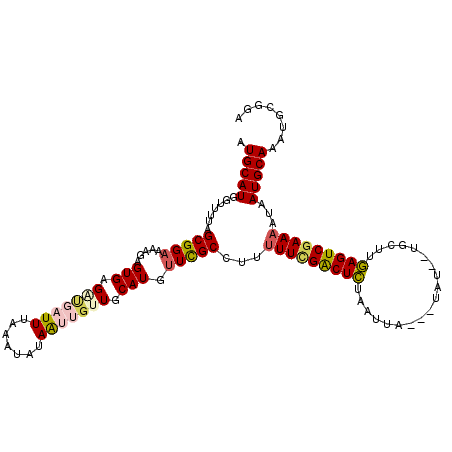
| Location | 12,604,129 – 12,604,243 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 86.65 |
| Mean single sequence MFE | -25.82 |
| Consensus MFE | -18.55 |
| Energy contribution | -19.15 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12604129 114 - 23771897 AUGCAUGGUUUUAGCGGAAAAGAGUGAGAUGAUUUAAAUAUAAUUGUUGCAUGUUCGCCUUUUUCGACUCUAAUUACUAUAUAUUGCUUGAGUCGAAAAUAAUGCAAAUGCGGA .(((((.(((((...(((((((.((((((((.....((((....)))).))).))))))))))))(((((...................))))).))))).)))))........ ( -26.61) >DroSec_CAF1 92006 111 - 1 AUGCAUGGUUUUAGCGGAAAAGAGUGCGAUGAUUUAAAUAUAAUUGUUGCAUGUUCGCCUUUUUCGACUCUAAUUA---UAUACUUCUCGAGUCAAAAAUAAUGCAAAUGCGGA .(((((.(((((.(((((.....((((((..(((.......)))..)))))).))))).......(((((......---..........))))).))))).)))))........ ( -28.49) >DroSim_CAF1 94815 109 - 1 AUGCAUGGUUUUAGCGGAAAAGAGUGAGUAGAUUUAAAUAUAAUUGUUGCAUGUUCGCCUUUUUCGACUCUAAUUA---UAU--UUCUCGAGUCGAAAAUAAUGCAAAUGCGGA .(((((.(((((...(((((((.(((((((......((((....))))...))))))))))))))(((((......---...--.....))))).))))).)))))........ ( -24.74) >DroEre_CAF1 95267 107 - 1 AUGCAUGGUUUAAGCGGAAUUGAGUGAGGCGAUUUAAAUAUAGAAGUUGCAUGUUCGCCU--UUUGGCUUUAAUUA---UAU--UGCUUAAGUCGAAAAUAAUGCAAAUACGGA .(((((.((((...(((..(((((..(.(((((((........)))))))......(((.--...)))........---..)--..))))).))).)))).)))))........ ( -24.70) >DroYak_CAF1 95664 109 - 1 UUGCAUGGCUUAAGCGGAAUAGAGUGAGUUUAUUUAAAUAUAGAUGUUGCAUGCUUGCCUAUUUUGACUUUAAUUA---UAU--UGCUUAAGUCGAAAAUAAUGCAAAUGCGGA ((((((((((((((((((((((.(..(((.((((((....))))))......)))..)))))))............---...--)))))))))).......))))))....... ( -24.57) >consensus AUGCAUGGUUUUAGCGGAAAAGAGUGAGAUGAUUUAAAUAUAAUUGUUGCAUGUUCGCCUUUUUCGACUCUAAUUA___UAU__UGCUUGAGUCGAAAAUAAUGCAAAUGCGGA .(((((.......(((((.....(((.(((((((.......))))))).))).)))))...(((((((((...................)))))))))...)))))........ (-18.55 = -19.15 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:51 2006