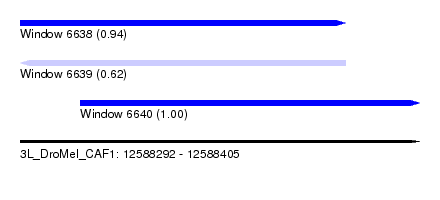
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,588,292 – 12,588,405 |
| Length | 113 |
| Max. P | 0.998860 |

| Location | 12,588,292 – 12,588,384 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 73.96 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -11.66 |
| Energy contribution | -12.30 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12588292 92 + 23771897 UUGUCUUAGUGGUGAUUGCCUGCCCUUCAAAUGGUUUCUCACUGGCAAUUAGUGGGAAACACAUUCUGGCAAUAUUGAUGACUGGUUUUUGC ..(((((((((((....)))((((.....(((((((((((((((.....))))))))))).))))..))))..))))).))).......... ( -26.50) >DroSec_CAF1 76367 86 + 1 UUGUCUUACUGGUGAUUGAAGGGCCUUUAGAUGGUUUCUCACUGGUAAUUAGUGGGAAACACAUUCUGGCAAUAUUGAUGACU------UGU .......((.(((.((..(...(((....(((((((((((((((.....))))))))))).))))..)))....)..)).)))------.)) ( -26.70) >DroSim_CAF1 79150 86 + 1 UCUUCUCACUGGCGAUUGAAGGGCCUUCAAAUUGUUUCUCACUGGUAAUUAGUGGGAAACACAUUCUGGCAAUAUUGAUGACU------UGC (((((............)))))(((.......((((((((((((.....))))))))))))......))).............------... ( -21.52) >DroEre_CAF1 79063 92 + 1 UCGAUUCACUGCAGAUUGCCUGCCCUUCAGAUGGAUUCGUACUGGCCAUUAGUGGGCAGCACAUUCUGGCCAUAUUGAUGGCUAGCUUUCGU .(((((((((((((.....)))).....)).)))).)))..((((((((((((((.(((......))).))))..))))))))))....... ( -29.90) >DroYak_CAF1 78704 76 + 1 UCGAUUCAC----------------UUCAGAUGGAUUCUUGCUGGCAAUUAGUGGGAAACACAUUCUGGCCAUAUUGAUGGCUAGUUUCCGU ..((.....----------------.))..(((((((((..(((.....)))..)))).......((((((((....))))))))..))))) ( -22.40) >consensus UCGUCUCACUGGUGAUUGAAGGCCCUUCAGAUGGUUUCUCACUGGCAAUUAGUGGGAAACACAUUCUGGCAAUAUUGAUGACU_G_UU_UGU ..........................(((((..(((((((((((.....)))))))))))....)))))....................... (-11.66 = -12.30 + 0.64)



| Location | 12,588,292 – 12,588,384 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 73.96 |
| Mean single sequence MFE | -19.84 |
| Consensus MFE | -6.02 |
| Energy contribution | -7.38 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.30 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12588292 92 - 23771897 GCAAAAACCAGUCAUCAAUAUUGCCAGAAUGUGUUUCCCACUAAUUGCCAGUGAGAAACCAUUUGAAGGGCAGGCAAUCACCACUAAGACAA ..........(((........((((.(((((.(((((.((((.......)))).))))))))))....))))((......)).....))).. ( -24.00) >DroSec_CAF1 76367 86 - 1 ACA------AGUCAUCAAUAUUGCCAGAAUGUGUUUCCCACUAAUUACCAGUGAGAAACCAUCUAAAGGCCCUUCAAUCACCAGUAAGACAA ...------.(((.........((((((.((.(((((.((((.......)))).))))))))))...))).((.........))...))).. ( -14.60) >DroSim_CAF1 79150 86 - 1 GCA------AGUCAUCAAUAUUGCCAGAAUGUGUUUCCCACUAAUUACCAGUGAGAAACAAUUUGAAGGCCCUUCAAUCGCCAGUGAGAAGA ...------.............(((.((((.((((((.((((.......)))).))))))))))...))).((((..(((....))))))). ( -17.80) >DroEre_CAF1 79063 92 - 1 ACGAAAGCUAGCCAUCAAUAUGGCCAGAAUGUGCUGCCCACUAAUGGCCAGUACGAAUCCAUCUGAAGGGCAGGCAAUCUGCAGUGAAUCGA .(((..(((.(((((....)))))((((.(((.((((((.(..((((...........))))..)..))))))))).)))).)))...))). ( -29.10) >DroYak_CAF1 78704 76 - 1 ACGGAAACUAGCCAUCAAUAUGGCCAGAAUGUGUUUCCCACUAAUUGCCAGCAAGAAUCCAUCUGAA----------------GUGAAUCGA .(((..(((.(((((....)))))((((..(((.....)))...(((....))).......)))).)----------------))...))). ( -13.70) >consensus ACA_AA_C_AGUCAUCAAUAUUGCCAGAAUGUGUUUCCCACUAAUUGCCAGUGAGAAACCAUCUGAAGGCCAGGCAAUCACCAGUGAGACGA ........................((((.((.(((((.((((.......)))).)))))))))))........................... ( -6.02 = -7.38 + 1.36)



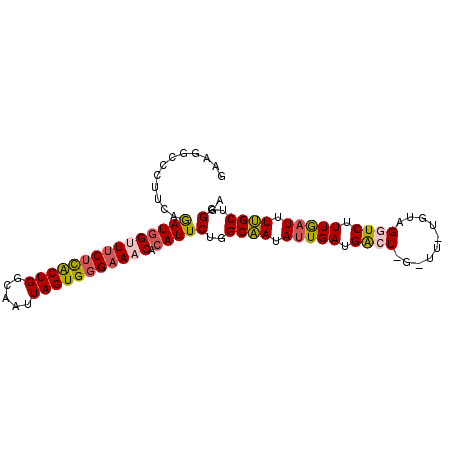
| Location | 12,588,309 – 12,588,405 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 78.72 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -22.82 |
| Energy contribution | -22.90 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.72 |
| SVM decision value | 3.26 |
| SVM RNA-class probability | 0.998860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12588309 96 + 23771897 GCCUGCCCUUCAAAUGGUUUCUCACUGGCAAUUAGUGGGAAACACAUUCUGGCAAUAUUGAUGACUGGUUUUUGCAGGUCUUUGAUUUUGCUGGGU ....((((....(((((((((((((((.....))))))))))).))))..(((((.((..(.((((.((....)).)))).)..)).))))))))) ( -34.30) >DroSec_CAF1 76384 90 + 1 GAAGGGCCUUUAGAUGGUUUCUCACUGGUAAUUAGUGGGAAACACAUUCUGGCAAUAUUGAUGACU------UGUAGGUCUUUGAUUUUGCUGGGC .....((((...(((((((((((((((.....))))))))))).))))..(((((.((..(.((((------....)))).)..)).))))))))) ( -31.70) >DroSim_CAF1 79167 90 + 1 GAAGGGCCUUCAAAUUGUUUCUCACUGGUAAUUAGUGGGAAACACAUUCUGGCAAUAUUGAUGACU------UGCGGGUCUUUGAUUUUGCUGGGU ((((...))))....((((((((((((.....))))))))))))...((..((((.((..(.((((------....)))).)..)).))))..)). ( -31.00) >DroEre_CAF1 79080 96 + 1 GCCUGCCCUUCAGAUGGAUUCGUACUGGCCAUUAGUGGGCAGCACAUUCUGGCCAUAUUGAUGGCUAGCUUUCGUAGUUCUUUAAUUUGGCUGGAA .((.(((........(((((((..((((((((((((((.(((......))).))))..))))))))))....)).)))))........))).)).. ( -33.89) >DroYak_CAF1 78713 88 + 1 --------UUCAGAUGGAUUCUUGCUGGCAAUUAGUGGGAAACACAUUCUGGCCAUAUUGAUGGCUAGUUUCCGUAGCUCUUUGUUUUGGCUGGGA --------....(((((.((((..(((.....)))..)))).).))))((((((((....))))))))..(((.((((.(........)))))))) ( -26.50) >consensus GAAGGCCCUUCAGAUGGUUUCUCACUGGCAAUUAGUGGGAAACACAUUCUGGCAAUAUUGAUGACU_G_UU_UGUAGGUCUUUGAUUUUGCUGGGA ............(((((((((((((((.....))))))))))).))))(..((((.(((((.((((..........)))).))))).))))..).. (-22.82 = -22.90 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:47 2006