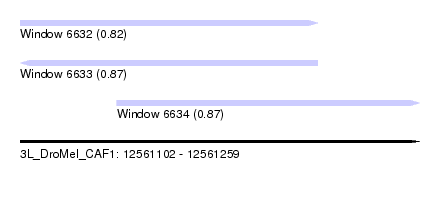
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,561,102 – 12,561,259 |
| Length | 157 |
| Max. P | 0.865777 |

| Location | 12,561,102 – 12,561,219 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.02 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -22.24 |
| Energy contribution | -22.80 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12561102 117 + 23771897 AGUGUGUGUGAGCGCCCACACAGUGGGGGGUAGAAAUA---GUAAUAGCAGUACUUUGCCUGCAAAAGCUAUUUAUUAUAAUAGCAAAGCAGUUUCAGGAUAUCCUGAACAACAAUUUAA .(((((.((....)).))))).((..(((((((((((.---((....((((........))))....((((((......))))))...)).)))))....))))))..)).......... ( -30.50) >DroSec_CAF1 49288 117 + 1 UGUGUGUGUGAGCGCCCACACAGUGGGGGGCAGAAAUA---GUAAUAGCAGUACUUUGCCUGCAAAAGCUAUUUAUUAUAAUAGCAAAACAUUUUCAGGAUAUCCUGAACAACAAUUUAA ((((((.((....)).))))))((((.((((((....(---(((.......)))))))))).)....((((((......))))))....))).((((((....))))))........... ( -32.40) >DroSim_CAF1 50682 117 + 1 UGUGUGUGUGAGCGCCCACACAGUGAGGGGCAGAAAUA---GUAAUAGCAGUACUUUGCCUGCAAAAGCUAUUUAUUAUAAUAGCAAAACAGUUUCAGGAUAUCCUAAACAACAAUUUAA ..(((.(((..((.(((((...))).)).)).(((((.---((....((((........))))....((((((......))))))...)).)))))(((....)))..))))))...... ( -28.00) >DroEre_CAF1 50000 110 + 1 ----UGUGUGAGCGCCCACACAGUGGGGGGCAGAAAUA---GUAAUAGCAGUACUUUGGCUGCCAAAGCUAUUUAUUAUACUAGCAAAACUGUUUGAGGAUAUCCUGAACAAUUUUU--- ----.((((((((.(((........))).))..(((((---((....(((((......)))))....))))))).))))))....((((.(((((.(((....)))))))).)))).--- ( -31.70) >DroYak_CAF1 50666 113 + 1 ----UGUGUGAGCGCCCACACAGUGGGGGGCAGUAAUAAUAAUAAUAGCAGUACUUUGUCUGCAAAACCUAUUUAUUAUAAUAGCAAAACAGUUUUAGGAUAUCCUGAACAAGAAUU--- ----..(((..((((((.(.....)..)))).(((((((........((((........)))).........)))))))....))...)))((((.(((....))))))).......--- ( -24.13) >consensus _GUGUGUGUGAGCGCCCACACAGUGGGGGGCAGAAAUA___GUAAUAGCAGUACUUUGCCUGCAAAAGCUAUUUAUUAUAAUAGCAAAACAGUUUCAGGAUAUCCUGAACAACAAUUUAA ...........((.(((((...))).)).))................((((........))))....((((((......))))))........((((((....))))))........... (-22.24 = -22.80 + 0.56)


| Location | 12,561,102 – 12,561,219 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.02 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -21.20 |
| Energy contribution | -21.92 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12561102 117 - 23771897 UUAAAUUGUUGUUCAGGAUAUCCUGAAACUGCUUUGCUAUUAUAAUAAAUAGCUUUUGCAGGCAAAGUACUGCUAUUAC---UAUUUCUACCCCCCACUGUGUGGGCGCUCACACACACU .....((((..((((((....)))))).((((...((((((......))))))....))))))))((((.......)))---)...............(((((((....))))))).... ( -29.50) >DroSec_CAF1 49288 117 - 1 UUAAAUUGUUGUUCAGGAUAUCCUGAAAAUGUUUUGCUAUUAUAAUAAAUAGCUUUUGCAGGCAAAGUACUGCUAUUAC---UAUUUCUGCCCCCCACUGUGUGGGCGCUCACACACACA ...........((((((....))))))..(((...((((((......))))))....)))(((((((((.((....)).---))))).))))......(((((((....))))))).... ( -27.70) >DroSim_CAF1 50682 117 - 1 UUAAAUUGUUGUUUAGGAUAUCCUGAAACUGUUUUGCUAUUAUAAUAAAUAGCUUUUGCAGGCAAAGUACUGCUAUUAC---UAUUUCUGCCCCUCACUGUGUGGGCGCUCACACACACA ...........((((((....)))))).((((...((((((......))))))....))))((..((((.......)))---)......(((((.......).))))))........... ( -25.70) >DroEre_CAF1 50000 110 - 1 ---AAAAAUUGUUCAGGAUAUCCUCAAACAGUUUUGCUAGUAUAAUAAAUAGCUUUGGCAGCCAAAGUACUGCUAUUAC---UAUUUCUGCCCCCCACUGUGUGGGCGCUCACACA---- ---.(((((((((.(((....)))..)))))))))((((..........))))...(((((....((((.......)))---)....))))).......((((((....)))))).---- ( -27.10) >DroYak_CAF1 50666 113 - 1 ---AAUUCUUGUUCAGGAUAUCCUAAAACUGUUUUGCUAUUAUAAUAAAUAGGUUUUGCAGACAAAGUACUGCUAUUAUUAUUAUUACUGCCCCCCACUGUGUGGGCGCUCACACA---- ---...........(((....)))...........((....(((((((.(((.....((((........))))..))))))))))....))........((((((....)))))).---- ( -18.60) >consensus UUAAAUUGUUGUUCAGGAUAUCCUGAAACUGUUUUGCUAUUAUAAUAAAUAGCUUUUGCAGGCAAAGUACUGCUAUUAC___UAUUUCUGCCCCCCACUGUGUGGGCGCUCACACACAC_ ...........((((((....))))))........((((((......))))))....((((........)))).........................(((((((....))))))).... (-21.20 = -21.92 + 0.72)

| Location | 12,561,140 – 12,561,259 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 86.83 |
| Mean single sequence MFE | -20.81 |
| Consensus MFE | -15.07 |
| Energy contribution | -15.82 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12561140 119 + 23771897 GUAAUAGCAGUACUUUGCCUGCAAAAGCUAUUUAUUAUAAUAGCAAAGCAGUUUCAGGAUAUCCUGAACAACAAUUUAAAAAUUUCAACAGCUUAGUAAUUUAUAAAAUAAGAUAAUAC ...((((...((((..((((((....((((((......))))))...)))).((((((....))))))......................))..))))..))))............... ( -23.50) >DroSec_CAF1 49326 119 + 1 GUAAUAGCAGUACUUUGCCUGCAAAAGCUAUUUAUUAUAAUAGCAAAACAUUUUCAGGAUAUCCUGAACAACAAUUUAAAAAUUUCAAUAACUCAGUAAUUUAUAAAAUAAGAGAACAU ......((((........))))....((((((......))))))........((((((....)))))).......................(((.................)))..... ( -19.03) >DroSim_CAF1 50720 119 + 1 GUAAUAGCAGUACUUUGCCUGCAAAAGCUAUUUAUUAUAAUAGCAAAACAGUUUCAGGAUAUCCUAAACAACAAUUUAAAAAUUUCAACAGCUUAGUACUUUAAUAAAUAAGAGAAUAU ....(((.((((((..((.((.....((((((......))))))......((((.(((....)))))))...................))))..)))))))))................ ( -21.10) >DroEre_CAF1 50034 114 + 1 GUAAUAGCAGUACUUUGGCUGCCAAAGCUAUUUAUUAUACUAGCAAAACUGUUUGAGGAUAUCCUGAACAAUUUUU---AAAUCUCCACAGCUAAGUAAUUUAUAAAA--AUUUCACAU ...((((...(((((.(((((.....((((..........))))((((.(((((.(((....)))))))).)))).---.........))))))))))..))))....--......... ( -19.60) >consensus GUAAUAGCAGUACUUUGCCUGCAAAAGCUAUUUAUUAUAAUAGCAAAACAGUUUCAGGAUAUCCUGAACAACAAUUUAAAAAUUUCAACAGCUUAGUAAUUUAUAAAAUAAGAGAACAU ......((((........))))....((((((......))))))........((((((....))))))................................................... (-15.07 = -15.82 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:41 2006