
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,557,629 – 12,557,813 |
| Length | 184 |
| Max. P | 0.998944 |

| Location | 12,557,629 – 12,557,722 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 91.51 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -26.02 |
| Energy contribution | -26.58 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12557629 93 + 23771897 GUCCAACGGGCGGAGCACUCAACUUAAUUAUGGGUUCGGUGCGUCGAUAUUUUCGCCCAGAAGACUAAAGUCUUCAGAUUCCAGACCUUAAGA (((....(((((..(((((.((((((....)))))).)))))..)((.....)))))).((((((....))))))........)))....... ( -31.90) >DroSec_CAF1 45866 93 + 1 GUCCAACGGGCGGAGCACUCAACUUAAUUAUGGGUUCGGUGCGUCGAUAUUUUCGCCCAGAAGACUAAAGUCUUCAGAUUCCAGACUUCAGAA (((....(((((..(((((.((((((....)))))).)))))..)((.....)))))).((((((....))))))........)))....... ( -32.30) >DroSim_CAF1 47112 93 + 1 GUCCAACGGGCGGAGCACUCAACUUAAUUAUGGGUUCGGUGCGUCAAUAUUUUCGCCCAGAAGACUAAAGUCCUCAGAUUCCAGACUUCAGAA (((....((((((((((((.((((((....)))))).))))).........))))))).((.(((....))).))........)))....... ( -27.50) >DroEre_CAF1 46583 93 + 1 GUCCAUCAGGCGGAGCACUCAACUUAAUUAUGGGUUCGGUGCUCCGAUAUUUUCGCCCAGAAGACUGAAGUCUUUAGAUUCCAGACUUCAGCA ........(((((((((((.((((((....)))))).))))))))((.....))))).....(.(((((((((.........)))))))))). ( -35.80) >DroYak_CAF1 47079 93 + 1 GUCCAUCAGGCGGAGCACUCAACUUAAUUAUGGGUUCGAUGCGCCGAUAUUUUCGCCCAGAAGACCGACGUCUUUAGAUUCCAGACUUGAAGA (((.(((.(((((.(((.((((((((....)))))).))))).))((.....)))))..((((((....)))))).)))....)))....... ( -22.60) >consensus GUCCAACGGGCGGAGCACUCAACUUAAUUAUGGGUUCGGUGCGUCGAUAUUUUCGCCCAGAAGACUAAAGUCUUCAGAUUCCAGACUUCAGAA (((....((((((((((((.((((((....)))))).))))).........))))))).((((((....))))))........)))....... (-26.02 = -26.58 + 0.56)



| Location | 12,557,629 – 12,557,722 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 91.51 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -24.76 |
| Energy contribution | -23.84 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12557629 93 - 23771897 UCUUAAGGUCUGGAAUCUGAAGACUUUAGUCUUCUGGGCGAAAAUAUCGACGCACCGAACCCAUAAUUAAGUUGAGUGCUCCGCCCGUUGGAC ...((((((((.........))))))))((((..((((((......(((((...................)))))......))))))..)))) ( -26.21) >DroSec_CAF1 45866 93 - 1 UUCUGAAGUCUGGAAUCUGAAGACUUUAGUCUUCUGGGCGAAAAUAUCGACGCACCGAACCCAUAAUUAAGUUGAGUGCUCCGCCCGUUGGAC ..(((((((((.........)))))))))(((..((((((......(((((...................)))))......))))))..))). ( -26.71) >DroSim_CAF1 47112 93 - 1 UUCUGAAGUCUGGAAUCUGAGGACUUUAGUCUUCUGGGCGAAAAUAUUGACGCACCGAACCCAUAAUUAAGUUGAGUGCUCCGCCCGUUGGAC ..(((((((((.........)))))))))(((..((((((........((.((((..(((..........)))..))))))))))))..))). ( -26.10) >DroEre_CAF1 46583 93 - 1 UGCUGAAGUCUGGAAUCUAAAGACUUCAGUCUUCUGGGCGAAAAUAUCGGAGCACCGAACCCAUAAUUAAGUUGAGUGCUCCGCCUGAUGGAC .((((((((((.........))))))))))((..(((((((.....))(((((((..(((..........)))..))))))))))))..)).. ( -33.90) >DroYak_CAF1 47079 93 - 1 UCUUCAAGUCUGGAAUCUAAAGACGUCGGUCUUCUGGGCGAAAAUAUCGGCGCAUCGAACCCAUAAUUAAGUUGAGUGCUCCGCCUGAUGGAC .......((((.........))))(((.(((....((((((.....))((.((((..(((..........)))..)))).))))))))).))) ( -22.60) >consensus UCCUGAAGUCUGGAAUCUGAAGACUUUAGUCUUCUGGGCGAAAAUAUCGACGCACCGAACCCAUAAUUAAGUUGAGUGCUCCGCCCGUUGGAC ...((((((((.........))))))))((((..((((((........((.((((..(((..........)))..))))))))))))..)))) (-24.76 = -23.84 + -0.92)

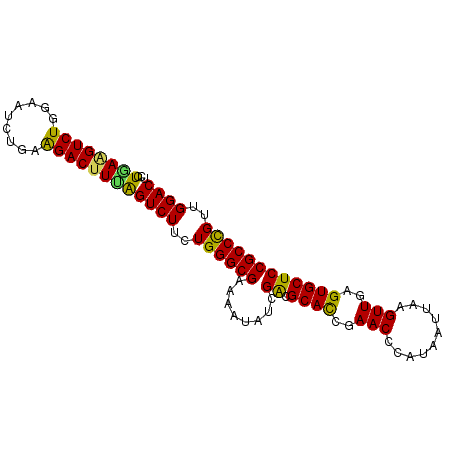

| Location | 12,557,659 – 12,557,761 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 80.91 |
| Mean single sequence MFE | -28.26 |
| Consensus MFE | -21.30 |
| Energy contribution | -22.02 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12557659 102 - 23771897 GUUUGAAGUCUGGAAGUCUUGAAGUUUG-AAGUCUGGAAGUCUUAAGGUCUGGAAUCUGAAGACUUUAGUCUUCUGGGCGAAAAUAUCGACGCACCGAACCCA .......((.(((..((.((((.((((.-..(((..((((.((.(((((((.........))))))))).))))..)))..)))).)))).)).))).))... ( -29.70) >DroSec_CAF1 45896 102 - 1 ACUUGAAGUCUGGAAGUCUUGAAGCUUG-AAGUCUGGAAGUUCUGAAGUCUGGAAUCUGAAGACUUUAGUCUUCUGGGCGAAAAUAUCGACGCACCGAACCCA .(((.(((.(.....).))).)))....-..(((..((((..(((((((((.........))))))))).))))..))).......(((......)))..... ( -29.00) >DroSim_CAF1 47142 93 - 1 ---------CUGGAAGUCUUGAAGCUUG-AAGUCUGGAAGUUCUGAAGUCUGGAAUCUGAGGACUUUAGUCUUCUGGGCGAAAAUAUUGACGCACCGAACCCA ---------.(((..(((..........-..(((..((((..(((((((((.........))))))))).))))..))).........)))...)))...... ( -26.40) >DroEre_CAF1 46613 87 - 1 GU----------------UUGAAGUCUGGAAGUCCUGAAGUGCUGAAGUCUGGAAUCUAAAGACUUCAGUCUUCUGGGCGAAAAUAUCGGAGCACCGAACCCA ..----------------.........((..((((.((((.((((((((((.........)))))))))))))).)))).......((((....))))..)). ( -31.70) >DroYak_CAF1 47109 96 - 1 GU-------CUGGAAGUCUUGAAGUCUGUAAGUCUUAAAGUCUUCAAGUCUGGAAUCUAAAGACGUCGGUCUUCUGGGCGAAAAUAUCGGCGCAUCGAACCCA ((-------(..((((.(((((((.((...........)).))))))).((((..((....))..)))).))))..))).......((((....))))..... ( -24.50) >consensus GU_______CUGGAAGUCUUGAAGUUUG_AAGUCUGGAAGUCCUGAAGUCUGGAAUCUGAAGACUUUAGUCUUCUGGGCGAAAAUAUCGACGCACCGAACCCA ...............((.((((.((((....(((((((((..(((((((((.........))))))))).)))))))))..)))).)))).)).......... (-21.30 = -22.02 + 0.72)


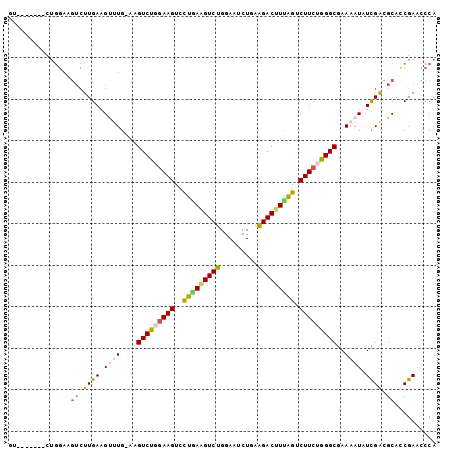
| Location | 12,557,722 – 12,557,813 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 86.08 |
| Mean single sequence MFE | -24.17 |
| Consensus MFE | -19.82 |
| Energy contribution | -20.48 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12557722 91 + 23771897 CUUCCAGACUUCAAACUUCAAGACUUCCAGACUUCAAACUUCCAGACUUCAGUUUGCAGAUUGCAGACUGUGGGGAAAUUGAAGCAGUCCG ......((((.....((((((...((((.(....)...........((.((((((((.....)))))))).)))))).)))))).)))).. ( -23.30) >DroSec_CAF1 45959 91 + 1 CUUCCAGACUUCAAGCUUCAAGACUUCCAGACUUCAAGUUUCCAGACUUCAGUUUACAGAUUGCAGACUGUGAGGAAAUUGAAGCAGUCCG ......((((....(((((((...((((..(((..(((((....))))).)))((((((........)))))))))).))))))))))).. ( -23.40) >DroSim_CAF1 47205 76 + 1 CUUCCAGACUUCAAGCUUCAAGACUUCCAG---------------ACUUCAGUUUGCAGAUUGCAGACUGUGAGGAAAUUGAAGCAGUCCG ......((((....(((((((...((((..---------------....((((((((.....))))))))...)))).))))))))))).. ( -25.80) >consensus CUUCCAGACUUCAAGCUUCAAGACUUCCAGACUUCAA__UUCCAGACUUCAGUUUGCAGAUUGCAGACUGUGAGGAAAUUGAAGCAGUCCG ......((((....(((((((...((((.....................((((((((.....))))))))...)))).))))))))))).. (-19.82 = -20.48 + 0.67)


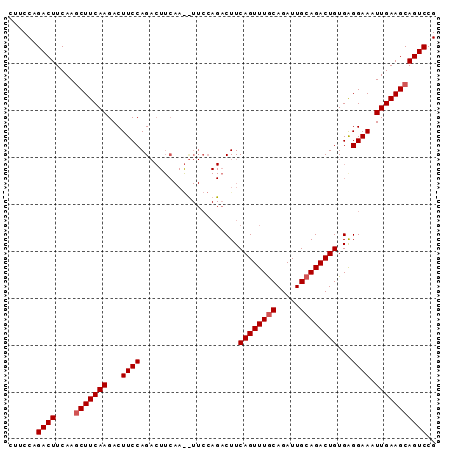
| Location | 12,557,722 – 12,557,813 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 86.08 |
| Mean single sequence MFE | -26.57 |
| Consensus MFE | -25.24 |
| Energy contribution | -24.97 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.984102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12557722 91 - 23771897 CGGACUGCUUCAAUUUCCCCACAGUCUGCAAUCUGCAAACUGAAGUCUGGAAGUUUGAAGUCUGGAAGUCUUGAAGUUUGAAGUCUGGAAG ((((((((((((((((((...((((.(((.....))).)))).((.((..........)).))))))))..))))))....)))))).... ( -26.20) >DroSec_CAF1 45959 91 - 1 CGGACUGCUUCAAUUUCCUCACAGUCUGCAAUCUGUAAACUGAAGUCUGGAAACUUGAAGUCUGGAAGUCUUGAAGCUUGAAGUCUGGAAG ((((((((((((((((((...((((.(((.....))).))))...((.(....)..)).....))))))..))))))....)))))).... ( -27.20) >DroSim_CAF1 47205 76 - 1 CGGACUGCUUCAAUUUCCUCACAGUCUGCAAUCUGCAAACUGAAGU---------------CUGGAAGUCUUGAAGCUUGAAGUCUGGAAG ((((((((((((((((((...((((.(((.....))).))))....---------------..))))))..))))))....)))))).... ( -26.30) >consensus CGGACUGCUUCAAUUUCCUCACAGUCUGCAAUCUGCAAACUGAAGUCUGGAA__UUGAAGUCUGGAAGUCUUGAAGCUUGAAGUCUGGAAG ((((((((((((((((((...((((.(((.....))).)))).((.((..........)).))))))))..))))))....)))))).... (-25.24 = -24.97 + -0.27)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:38 2006