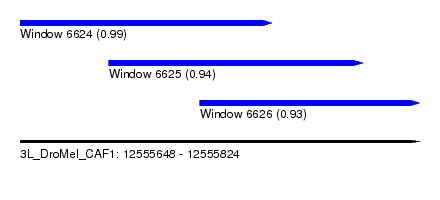
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,555,648 – 12,555,824 |
| Length | 176 |
| Max. P | 0.992944 |

| Location | 12,555,648 – 12,555,759 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 92.74 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -24.02 |
| Energy contribution | -23.86 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12555648 111 + 23771897 AGCGCUUUUGGACUUUUCACUAAGCAGCAAAAACAGCUGCUUGAGGCAAACUUUUUCCAUGUUUAUGACAGUAAAUUGGUGGCAGGCCAGCAA--------AAAACCAUAAUAAAUCAG ...(((..............((((((((.......)))))))).(((...((....(((.((((((....))))))))).))...))))))..--------.................. ( -24.30) >DroSec_CAF1 43892 116 + 1 AGCGCUUUUGGACUUUUCACUAAGCAGCAAAAACGGCUGCUUGAGGCAAACUUUUUCCAUGUUUAUGACAGUAAAUUGGUGGCAGGCCAGCAAA---AAAAAAAACCAUAAUAAAUCAG ...(((..............((((((((.......)))))))).(((...((....(((.((((((....))))))))).))...))))))...---...................... ( -24.20) >DroSim_CAF1 45115 119 + 1 AGCGCUUUUGGACUUUUCACUAAGCAGCAAAAACAGCUGCUUGAGGCAAACUUUUUCCAUGUUUAUGACAGUAAAUUGGUGGCAGGCCAGCAAAACAAAAAAAAACCAUAAUAAAUCAG ...(((..............((((((((.......)))))))).(((...((....(((.((((((....))))))))).))...))))))............................ ( -24.30) >DroEre_CAF1 44594 110 + 1 AGCGCUUUCUGACUUUUAACUAAGCAGCAAAAACAGCUGCUUG-GGCAAACUUUUUCCAUGUUUAUGACAGUAAAUUGGUAGAAAGCCAGCAA--------AAAAGCAUAAUAAAUCAG .(.((((((((...(((..(((((((((.......))))))))-)..)))......(((.((((((....)))))))))))))))))).((..--------....))............ ( -30.10) >DroYak_CAF1 45027 111 + 1 AGCGCUUUCGGACUUUUCACUAAGCAGCAAAAACAGCUGCUUGAGGCAAACUUUUUCCAUGUUUAUGACAGUAAAUUGGUGGAAGGCCAGCAA--------AAAAGCAUAAUAAAUCAG ...(((((............((((((((.......)))))))).(((...((((..(((.((((((....)))))))))..))))))).....--------.)))))............ ( -28.50) >consensus AGCGCUUUUGGACUUUUCACUAAGCAGCAAAAACAGCUGCUUGAGGCAAACUUUUUCCAUGUUUAUGACAGUAAAUUGGUGGCAGGCCAGCAA________AAAACCAUAAUAAAUCAG ...(((..............((((((((.......)))))))).(((...((....(((.((((((....))))))))).))...))))))............................ (-24.02 = -23.86 + -0.16)

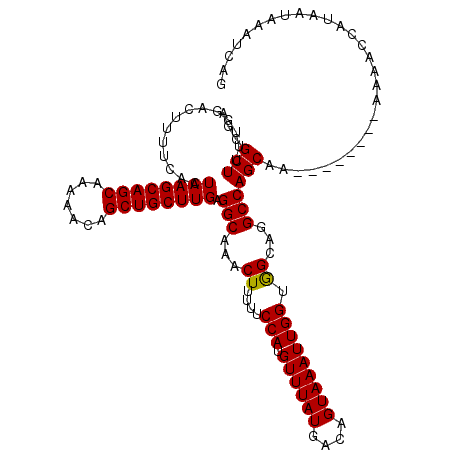

| Location | 12,555,687 – 12,555,799 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.80 |
| Mean single sequence MFE | -28.14 |
| Consensus MFE | -21.95 |
| Energy contribution | -22.35 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.940407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12555687 112 + 23771897 CUUGAGGCAAACUUUUUCCAUGUUUAUGACAGUAAAUUGGUGGCAGGCCAGCAA--------AAAACCAUAAUAAAUCAGAUGGCUGAUAAAUGGCCUGAGAAAAAGCCACUCCGAGGAA ((((..(....((((((((((((((((....))))))..))).(((((((....--------....((((..........))))........))))))).)))))))...)..))))... ( -27.49) >DroSec_CAF1 43931 117 + 1 CUUGAGGCAAACUUUUUCCAUGUUUAUGACAGUAAAUUGGUGGCAGGCCAGCAAA---AAAAAAAACCAUAAUAAAUCAGAUGGCUGAUAAAUGGCCUGAGAAAAGGGCACUCCGAGGAA ((((((((...((((((((((((((((....))))))..))))(((((((.....---.................(((((....)))))...))))))).)))))).)).)).))))... ( -29.41) >DroSim_CAF1 45154 120 + 1 CUUGAGGCAAACUUUUUCCAUGUUUAUGACAGUAAAUUGGUGGCAGGCCAGCAAAACAAAAAAAAACCAUAAUAAAUCAGAUGGCUGAUAAAUGGCCUGAGAAAAAGGCACUCCGAGGAA ((((((((...((((((((((((((((....))))))..))).((((((((.....)..................(((((....)))))...))))))).))))))))).)).))))... ( -28.30) >DroEre_CAF1 44633 111 + 1 CUUG-GGCAAACUUUUUCCAUGUUUAUGACAGUAAAUUGGUAGAAAGCCAGCAA--------AAAAGCAUAAUAAAUCAGAUGGCUGAUAAAUGGCCUGAGAAAAAGGCACUCGAAGGAA ((..-(((...(((((((((.((((((....))))))))).)))))).((((..--------....)).......(((((....)))))...)))))..))..........((....)). ( -25.40) >DroYak_CAF1 45066 112 + 1 CUUGAGGCAAACUUUUUCCAUGUUUAUGACAGUAAAUUGGUGGAAGGCCAGCAA--------AAAAGCAUAAUAAAUCAGAUGGCUGAUAAAUGGCCUGAGAAAAAGGCACUCCAAGGAA ((((((((........(((((((((((....))))))..)))))((((((((..--------....)).......(((((....)))))...)))))).........)).)).))))... ( -30.10) >consensus CUUGAGGCAAACUUUUUCCAUGUUUAUGACAGUAAAUUGGUGGCAGGCCAGCAA________AAAACCAUAAUAAAUCAGAUGGCUGAUAAAUGGCCUGAGAAAAAGGCACUCCGAGGAA ((((.((....(((((((((.((((((....))))))))....(((((((.........................(((((....)))))...))))))).)))))))...)).))))... (-21.95 = -22.35 + 0.40)

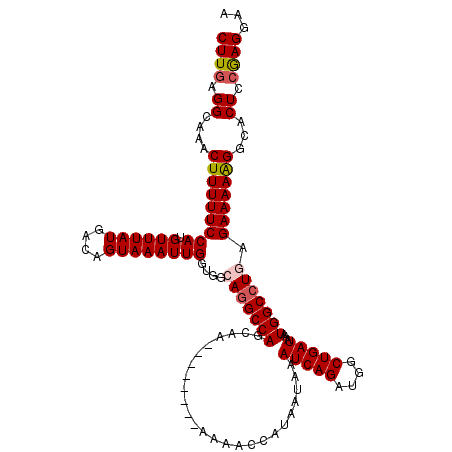
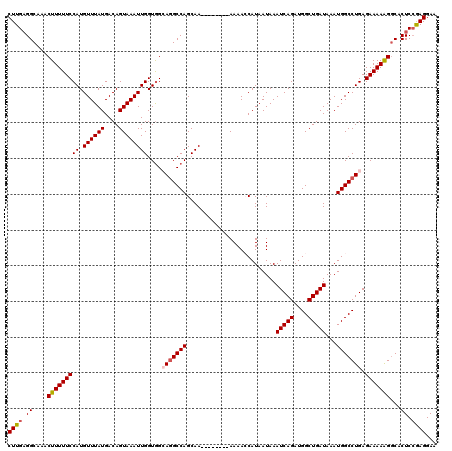
| Location | 12,555,727 – 12,555,824 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 91.35 |
| Mean single sequence MFE | -23.96 |
| Consensus MFE | -18.34 |
| Energy contribution | -18.98 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12555727 97 + 23771897 UGGCAGGCCAGCAA--------AAAACCAUAAUAAAUCAGAUGGCUGAUAAAUGGCCUGAGAAAAAGCCACUCCGAGGAAUCAGGCGUUUUUGUCAACCUUCAAA (((....)))....--------.................((.((.(((((((..((((((.......((.......))..))))))...))))))).)).))... ( -23.10) >DroSec_CAF1 43971 102 + 1 UGGCAGGCCAGCAAA---AAAAAAAACCAUAAUAAAUCAGAUGGCUGAUAAAUGGCCUGAGAAAAGGGCACUCCGAGGAAUCAGGCGUUUUUGUCAACCUUCAAA (((....))).....---.....................((.((.(((((((..((((((.....((.....))......))))))...))))))).)).))... ( -24.90) >DroSim_CAF1 45194 105 + 1 UGGCAGGCCAGCAAAACAAAAAAAAACCAUAAUAAAUCAGAUGGCUGAUAAAUGGCCUGAGAAAAAGGCACUCCGAGGAAUCAGGCGUUUUUGUCAACCUUCAAA (((....))).............................((.((.(((((((..((((((......((....))......))))))...))))))).)).))... ( -24.60) >DroEre_CAF1 44672 97 + 1 UAGAAAGCCAGCAA--------AAAAGCAUAAUAAAUCAGAUGGCUGAUAAAUGGCCUGAGAAAAAGGCACUCGAAGGAAUCAGGCGUUUUUGUCAACCUUCAAA ..(((.((((((..--------....)).......(((((....)))))...)))).(((.(((((.((..((....)).....)).))))).)))...)))... ( -23.00) >DroYak_CAF1 45106 97 + 1 UGGAAGGCCAGCAA--------AAAAGCAUAAUAAAUCAGAUGGCUGAUAAAUGGCCUGAGAAAAAGGCACUCCAAGGAAUCAGUCGUUUUUGUCAACCUUUAAA ..(((((...((..--------....))..........(((((((((((.....((((.......))))..((....))))))))))))).......)))))... ( -24.20) >consensus UGGCAGGCCAGCAA________AAAACCAUAAUAAAUCAGAUGGCUGAUAAAUGGCCUGAGAAAAAGGCACUCCGAGGAAUCAGGCGUUUUUGUCAACCUUCAAA (((....))).............................((.((.(((((((..((((((......((....))......))))))...))))))).)).))... (-18.34 = -18.98 + 0.64)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:34 2006