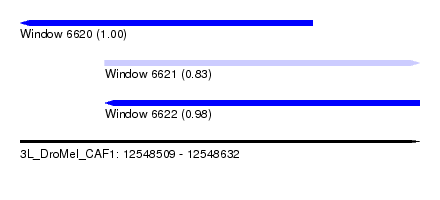
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,548,509 – 12,548,632 |
| Length | 123 |
| Max. P | 0.999367 |

| Location | 12,548,509 – 12,548,599 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 98.47 |
| Mean single sequence MFE | -33.30 |
| Consensus MFE | -32.00 |
| Energy contribution | -32.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.54 |
| SVM RNA-class probability | 0.999367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12548509 90 - 23771897 G---GGGGUGAUGAAAAGUAUCUGAGGGAUACACCAGUCAGUAUGCGCAGCUACAAGAUGGCUGCUGCCAAUCCUCGCCCGCACCAAUGCCAA (---.((((((.((...((((((((.((.....))..)))).))))(((((((.....)))))))......)).)))))).)........... ( -32.60) >DroSec_CAF1 36650 90 - 1 G---GGGGUGAUGAAAAGUAUCUGAGGGAUACACCAGUCAGUAUGCGCAGCUACAAGAUGGCUGCUGCCAAUCCUCGCCCGCACCAAUGCCAA (---.((((((.((...((((((((.((.....))..)))).))))(((((((.....)))))))......)).)))))).)........... ( -32.60) >DroSim_CAF1 37884 90 - 1 G---GGGGUGAUGAAAAGUAUCUGAGGGAUACACCAGUCAGUAUGCGCAGCUACAAGAUGGCUGCUGCCAAUCCUCGCCCGCACCAAUGCCAA (---.((((((.((...((((((((.((.....))..)))).))))(((((((.....)))))))......)).)))))).)........... ( -32.60) >DroEre_CAF1 37621 93 - 1 GGAGGGGGUGAUGAAAAGUAUCUGAGGGAUACACCAGUCAGUAUGCGCAGCUACAAGAUGGCUGCUGCCAAUCCUCGCCCGCACCAAUGCCAA ((.(.((((((.((...((((((((.((.....))..)))).))))(((((((.....)))))))......)).)))))).).))........ ( -35.50) >DroYak_CAF1 37621 91 - 1 GG--GGGGUGAUGAAAAGUAUCUGAGGGAUACACCAGUCAGUAUGCGCAGCUACAAGAUGGCUGCUGCCAAUCCUCGCCCGCACCAAUGCCAA ((--(((((((.((...((((((((.((.....))..)))).))))(((((((.....)))))))......)).)))))).).))........ ( -33.20) >consensus G___GGGGUGAUGAAAAGUAUCUGAGGGAUACACCAGUCAGUAUGCGCAGCUACAAGAUGGCUGCUGCCAAUCCUCGCCCGCACCAAUGCCAA .....((((((.((...((((((((.((.....))..)))).))))(((((((.....)))))))......)).))))))(((....)))... (-32.00 = -32.00 + 0.00)


| Location | 12,548,535 – 12,548,632 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 91.13 |
| Mean single sequence MFE | -15.50 |
| Consensus MFE | -13.40 |
| Energy contribution | -13.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12548535 97 + 23771897 CAGCAGCCAUCUUGUAGCUGCGCAUACUGACUGGUGUAUCCCUCAGAUACUUUUCAUCACCCC---CCUCAAUAUCCACUUCUUGGCU--UUU-UUUUUUCUA ..(((((.........)))))......(((.(((.(((((.....)))))...))))))....---.........(((.....)))..--...-......... ( -13.90) >DroSec_CAF1 36676 98 + 1 CAGCAGCCAUCUUGUAGCUGCGCAUACUGACUGGUGUAUCCCUCAGAUACUUUUCAUCACCCC---CCUCAAUAUCCACUUCUUGGCU--UUUAUUUUUUUUA ..(((((.........)))))......(((.(((.(((((.....)))))...))))))....---.........(((.....)))..--............. ( -13.90) >DroSim_CAF1 37910 98 + 1 CAGCAGCCAUCUUGUAGCUGCGCAUACUGACUGGUGUAUCCCUCAGAUACUUUUCAUCACCCC---CCUCAAUAUCCACUUCUUGGCU--UUUAUUUUUUUUA ..(((((.........)))))......(((.(((.(((((.....)))))...))))))....---.........(((.....)))..--............. ( -13.90) >DroEre_CAF1 37647 97 + 1 CAGCAGCCAUCUUGUAGCUGCGCAUACUGACUGGUGUAUCCCUCAGAUACUUUUCAUCACCCCCUCCCUCGAUAUCCAGCUCUUAGU------AUUUUUUUUA ..(((((.........)))))..(((((((((((.(((((.....))))).....(((............)))..))))...)))))------))........ ( -18.70) >DroYak_CAF1 37647 101 + 1 CAGCAGCCAUCUUGUAGCUGCGCAUACUGACUGGUGUAUCCCUCAGAUACUUUUCAUCACCCC--CCUUUAAAAUGCACUUCUUAGUUCGUUUUUUUUUUUUA ..(((((.........)))))......(((.(((.(((((.....)))))...))))))....--.....((((((.(((....))).))))))......... ( -17.10) >consensus CAGCAGCCAUCUUGUAGCUGCGCAUACUGACUGGUGUAUCCCUCAGAUACUUUUCAUCACCCC___CCUCAAUAUCCACUUCUUGGCU__UUUAUUUUUUUUA ..(((((.........)))))......(((.(((.(((((.....)))))...))))))............................................ (-13.40 = -13.40 + 0.00)


| Location | 12,548,535 – 12,548,632 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 91.13 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -19.00 |
| Energy contribution | -19.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12548535 97 - 23771897 UAGAAAAAA-AAA--AGCCAAGAAGUGGAUAUUGAGG---GGGGUGAUGAAAAGUAUCUGAGGGAUACACCAGUCAGUAUGCGCAGCUACAAGAUGGCUGCUG .........-...--...........(.(((((((..---((.....(....)(((((.....))))).))..))))))).)(((((((.....))))))).. ( -24.90) >DroSec_CAF1 36676 98 - 1 UAAAAAAAAUAAA--AGCCAAGAAGUGGAUAUUGAGG---GGGGUGAUGAAAAGUAUCUGAGGGAUACACCAGUCAGUAUGCGCAGCUACAAGAUGGCUGCUG .............--...........(.(((((((..---((.....(....)(((((.....))))).))..))))))).)(((((((.....))))))).. ( -24.90) >DroSim_CAF1 37910 98 - 1 UAAAAAAAAUAAA--AGCCAAGAAGUGGAUAUUGAGG---GGGGUGAUGAAAAGUAUCUGAGGGAUACACCAGUCAGUAUGCGCAGCUACAAGAUGGCUGCUG .............--...........(.(((((((..---((.....(....)(((((.....))))).))..))))))).)(((((((.....))))))).. ( -24.90) >DroEre_CAF1 37647 97 - 1 UAAAAAAAAU------ACUAAGAGCUGGAUAUCGAGGGAGGGGGUGAUGAAAAGUAUCUGAGGGAUACACCAGUCAGUAUGCGCAGCUACAAGAUGGCUGCUG ........((------(((....(((((.(((((..........)))))....(((((.....))))).))))).)))))..(((((((.....))))))).. ( -25.00) >DroYak_CAF1 37647 101 - 1 UAAAAAAAAAAAACGAACUAAGAAGUGCAUUUUAAAGG--GGGGUGAUGAAAAGUAUCUGAGGGAUACACCAGUCAGUAUGCGCAGCUACAAGAUGGCUGCUG ...........................((((((.....--)))))).......((((((((.((.....))..)))).))))(((((((.....))))))).. ( -20.30) >consensus UAAAAAAAAUAAA__AGCCAAGAAGUGGAUAUUGAGG___GGGGUGAUGAAAAGUAUCUGAGGGAUACACCAGUCAGUAUGCGCAGCUACAAGAUGGCUGCUG .....................................................((((((((.((.....))..)))).))))(((((((.....))))))).. (-19.00 = -19.00 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:30 2006