
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,541,854 – 12,541,955 |
| Length | 101 |
| Max. P | 0.774025 |

| Location | 12,541,854 – 12,541,955 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.77 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -21.83 |
| Energy contribution | -21.83 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
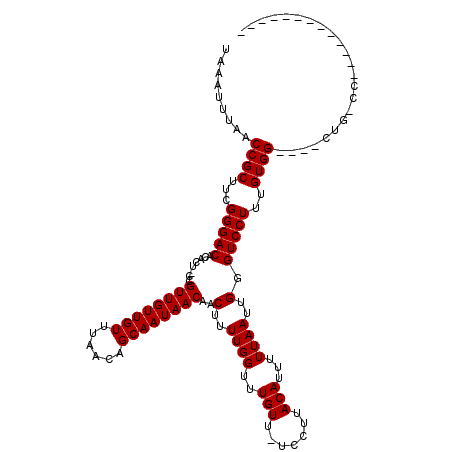
>3L_DroMel_CAF1 12541854 101 + 23771897 UAAAUUUAACCGCUUCGGGACACAGU-CCGUUGUUGUUUAACAGCAAUAACAACUUUUGGUUUGUU-UCCUUACAUUUUUAAUUGGGUCCUUUGUGG----CUGGCA------------- .........((((...(((((.((((-..((((((((......)))))))).......((......-.))...........)))).)))))..))))----......------------- ( -23.10) >DroPse_CAF1 29589 119 + 1 UAUAUUUAACCGCUUCGGGACACAGUUUCGUUGUUGUUUAACAGCAAUAACAACUUUUGGUUUGUU-UCCUUACAUUUUUAAUUGGGUCCUUUGUGGUCUUCUGGCCGCCCGAUGGCCAC ........(((((...(((((.(((((..((((((((......)))))))).......((......-.))..........))))).)))))..)))))....((((((.....)))))). ( -33.20) >DroGri_CAF1 32482 101 + 1 UAAAUUUAACCGCUUCGGGACACACU-GCGUUGUUGUUUAACAGCAAUAACAACUUUUGGUUUGUUUUCCUUACAUUUUUAAUUGGGUCCUUUGUGG----UCG-CC------------- ........(((((...(((((.....-..((((((((......))))))))..(..((((..(((.......)))...))))..).)))))..))))----)..-..------------- ( -23.30) >DroWil_CAF1 59526 100 + 1 UAAAUUUAACCGCUUCGGGACACACU-CCGUUGUUGUUUAACAGCAAUAACAUCUUUUGGUUUGUU-UCCUUACAUUUUUAAUUGGGUCCUUUGUGG----UUG-UU------------- ......(((((((...(((((.....-..((((((((......)))))))).((..((((..(((.-.....)))...))))..)))))))..))))----)))-..------------- ( -26.40) >DroMoj_CAF1 30740 100 + 1 UAAAUUUAACCGCUUCGGGACAUACG-GCGUUGUUGUUUAACAGCAAUAACAACUUUUGGUUUGUU-UCCUUACAUUUUUAAUUGGGUCCUUUGUGG----UCG-GU------------- .....(..(((((...(((((.....-..((((((((......))))))))..(..((((..(((.-.....)))...))))..).)))))..))))----)..-).------------- ( -24.70) >DroPer_CAF1 29591 119 + 1 UAUAUUUAACCGCUUCGGGACACAGUUUCGUUGUUGUUUAACAGCAAUAACAACUUUUGGUUUGUU-UCCUUACAUUUUUAAUUGGGUCCUUUGUGGUCUUCUGGCCGCCCGAUGGCCAC ........(((((...(((((.(((((..((((((((......)))))))).......((......-.))..........))))).)))))..)))))....((((((.....)))))). ( -33.20) >consensus UAAAUUUAACCGCUUCGGGACACACU_CCGUUGUUGUUUAACAGCAAUAACAACUUUUGGUUUGUU_UCCUUACAUUUUUAAUUGGGUCCUUUGUGG____CUG_CC_____________ .........((((...(((((........((((((((......))))))))..(..((((..(((.......)))...))))..).)))))..))))....................... (-21.83 = -21.83 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:25 2006