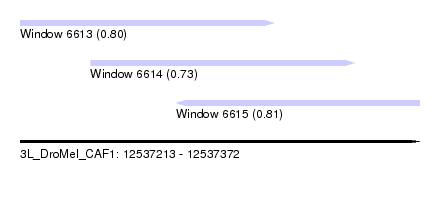
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,537,213 – 12,537,372 |
| Length | 159 |
| Max. P | 0.809713 |

| Location | 12,537,213 – 12,537,314 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 78.62 |
| Mean single sequence MFE | -26.16 |
| Consensus MFE | -17.27 |
| Energy contribution | -17.42 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.800632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12537213 101 + 23771897 CCUCAUGCCCA------UUCCCAUUC-CAGUUCCAAUUCGGA--AACGAAUCUCGAGAUGUCGCGAGCGUAGC-CAUAUUUCCAUGUCUCAUUUGUGGCGACCCCUCGAGA ...........------.........-.........((((..--..))))(((((((..(((((....)).((-((((...............)))))))))..))))))) ( -27.06) >DroPse_CAF1 25639 108 + 1 CCUCACCCUCCCCCCC-A-ACCGUGCUCAGCUCC-GCAGCAAUCUCUGAAUUUUAAGAUGUCGCGAGCGUAACUCAUUUUUCUAUGUCUCAUUUGGGGCGACCCCUCGAGA .(((........((((-(-(..(((....((((.-((.(((...(((........)))))).))))))......(((......)))...))))))))).((....))))). ( -22.20) >DroSec_CAF1 25364 107 + 1 CCUCAUGCUCAUGCCCAUACCCACUC-CAGUUCCAAUUCGGA--ACCCAAUCUCGAGAUGUCGCGAGCGUAGC-CAUAUUUCCAUGUCUCAUUUGCGGCGACCCCUCGAGA ......((....))............-..(((((.....)))--))....(((((((..(((((..(((.((.-(((......))).))....))).)))))..))))))) ( -29.00) >DroSim_CAF1 25906 106 + 1 CCUCAUGCUCAUGCCC-UACCCACUC-CAGUUCCAAUUCGGA--ACCCAAUCUCGAGAUGUCGCGAGCGUAGC-CAUAUUUCCAUGUCUCAUUUGCGGCGACCCCUCGAGA ......((....))..-.........-..(((((.....)))--))....(((((((..(((((..(((.((.-(((......))).))....))).)))))..))))))) ( -29.00) >DroEre_CAF1 26576 89 + 1 CCUCAUGC------CCUUUCCCA-------------UUCCAA--UCCGAAUCUCGAGAUGUCGCGAGCGGAGC-CAUAUUUCCAUGUCUCAUUUGCGGCGACCCCUCGAGA ........------.........-------------......--......(((((((..(((((..((.(((.-(((......))).)))....)).)))))..))))))) ( -25.50) >DroYak_CAF1 26091 107 + 1 CCUCCUGCCCCUGCCCUUUCCCAUUC-UCAGUCCAAUUCCAA--UCCGAAUCUGGAGAUGUCGCGAGCGGAGC-CAUAUUUCCAUGUCUCAUUUGCGGCGACCCCUCGAGA ........................((-((..((((((((...--...)))).))))..(((((((((..(((.-(((......))).))).))))))))).......)))) ( -24.20) >consensus CCUCAUGCUCAUGCCC_UUCCCAUUC_CAGUUCCAAUUCCAA__ACCGAAUCUCGAGAUGUCGCGAGCGUAGC_CAUAUUUCCAUGUCUCAUUUGCGGCGACCCCUCGAGA ..................................................(((((((..(((((..(((.....(((......))).......))).)))))..))))))) (-17.27 = -17.42 + 0.14)

| Location | 12,537,241 – 12,537,346 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 82.23 |
| Mean single sequence MFE | -29.28 |
| Consensus MFE | -20.28 |
| Energy contribution | -20.87 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12537241 105 + 23771897 AUUCGGA--AACGAAUCUCGAGAUGUCGCGAGCGUAGC-CAUAUUUCCAUGUCUCAUUUGUGGCGACCCCUCGAGAAUUGGAGCUCGCUC-UUUCCCCUAUAUAUUUUU----- .((((..--..))))(((((((..(((((....)).((-((((...............)))))))))..)))))))...((((....)))-).................----- ( -31.06) >DroPse_CAF1 25671 109 + 1 GCAGCAAUCUCUGAAUUUUAAGAUGUCGCGAGCGUAACUCAUUUUUCUAUGUCUCAUUUGGGGCGACCCCUCGAGAAUCGCUCCUCGUUUAUUUUCUUCUG-----UUUUUUUU ((.(((...(((........)))))).))(((((...(((..........(((((....)))))((....)))))...)))))..................-----........ ( -21.20) >DroSec_CAF1 25398 100 + 1 AUUCGGA--ACCCAAUCUCGAGAUGUCGCGAGCGUAGC-CAUAUUUCCAUGUCUCAUUUGCGGCGACCCCUCGAGAAUUGGAGCUCGCUC-UUUCCCCCAG-----UUU----- ....(((--(..((((((((((..(((((..(((.((.-(((......))).))....))).)))))..)))))).))))(((....)))-.)))).....-----...----- ( -30.60) >DroSim_CAF1 25939 100 + 1 AUUCGGA--ACCCAAUCUCGAGAUGUCGCGAGCGUAGC-CAUAUUUCCAUGUCUCAUUUGCGGCGACCCCUCGAGAAUUGGAGCUCGCUC-UUUCCCCCAG-----UUU----- ....(((--(..((((((((((..(((((..(((.((.-(((......))).))....))).)))))..)))))).))))(((....)))-.)))).....-----...----- ( -30.60) >DroEre_CAF1 26593 96 + 1 -UUCCAA--UCCGAAUCUCGAGAUGUCGCGAGCGGAGC-CAUAUUUCCAUGUCUCAUUUGCGGCGACCCCUCGAGAAUUGGAGCUCGCUC---UUCCCCAA-----UU------ -......--(((((.(((((((..(((((..((.(((.-(((......))).)))....)).)))))..))))))).)))))........---........-----..------ ( -33.00) >DroYak_CAF1 26125 100 + 1 AUUCCAA--UCCGAAUCUGGAGAUGUCGCGAGCGGAGC-CAUAUUUCCAUGUCUCAUUUGCGGCGACCCCUCGAGAAUUGGAGCUCGAUC-UUUUCCCCAA-----UUU----- .......--(((((.(((.(((..(((((..((.(((.-(((......))).)))....)).)))))..))).))).)))))........-..........-----...----- ( -29.20) >consensus AUUCCAA__ACCGAAUCUCGAGAUGUCGCGAGCGUAGC_CAUAUUUCCAUGUCUCAUUUGCGGCGACCCCUCGAGAAUUGGAGCUCGCUC_UUUCCCCCAG_____UUU_____ ..........((((.(((((((..(((((..(((.....(((......))).......))).)))))..))))))).))))................................. (-20.28 = -20.87 + 0.59)
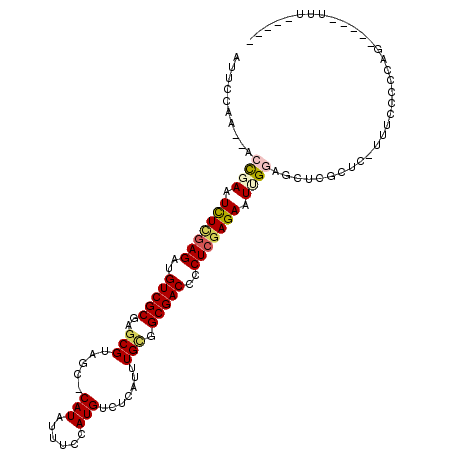
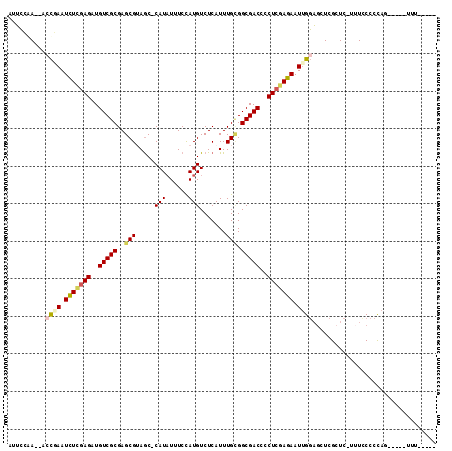
| Location | 12,537,275 – 12,537,372 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 85.32 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -24.37 |
| Energy contribution | -24.82 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12537275 97 - 23771897 GACGUGCGUGUGACCCCACUCAAAAA-------AAAAAUAUAUAGGGGAAAGAGC-GAGCUCCAAUUCUCGAGGGGUCGCCACAAAUGAGACAUGGAAAUAUGGC ..(((..((((((((((.((......-------..........))(((((.(((.-...)))...)))))..))))))).)))..)))...((((....)))).. ( -23.99) >DroSec_CAF1 25432 92 - 1 GACGUGCGUGUGACCCCACUCACAAA-------AAA-----CUGGGGGAAAGAGC-GAGCUCCAAUUCUCGAGGGGUCGCCGCAAAUGAGACAUGGAAAUAUGGC ....((((.((((((((.....((..-------...-----.)).(((((.(((.-...)))...)))))..)))))))))))).......((((....)))).. ( -30.40) >DroSim_CAF1 25973 92 - 1 GACGUGCGUGUGACCCCACUCACAAA-------AAA-----CUGGGGGAAAGAGC-GAGCUCCAAUUCUCGAGGGGUCGCCGCAAAUGAGACAUGGAAAUAUGGC ....((((.((((((((.....((..-------...-----.)).(((((.(((.-...)))...)))))..)))))))))))).......((((....)))).. ( -30.40) >DroEre_CAF1 26626 96 - 1 GACGUGCGUGUGACCCCACUCAAAAAACUCAAA-AA-----UUGGGGAA--GAGC-GAGCUCCAAUUCUCGAGGGGUCGCCGCAAAUGAGACAUGGAAAUAUGGC ....((((.((((((((.(((......((((..-..-----.))))...--)))(-(((........)))).)))))))))))).......((((....)))).. ( -31.00) >DroYak_CAF1 26159 99 - 1 GACGUGCGUGUGACCCCACUCAAAAACCUCAAAAAA-----UUGGGGAAAAGAUC-GAGCUCCAAUUCUCGAGGGGUCGCCGCAAAUGAGACAUGGAAAUAUGGC ....((((.((((((((.........((((((....-----))))))......((-(((........))))))))))))))))).......((((....)))).. ( -35.90) >DroAna_CAF1 22899 89 - 1 GACAUGCGUGCGACCCCACUCAAA----------UU-----UUGGGGAA-AAUGCAAAGCCCCAAUUCUCGAGGGGUCGCCGCAAAUGAGACAUGGAAAAAUUGU ....((((.((((((((..((...----------..-----((((((..-.........)))))).....))))))))))))))..................... ( -32.50) >consensus GACGUGCGUGUGACCCCACUCAAAAA_______AAA_____UUGGGGAAAAGAGC_GAGCUCCAAUUCUCGAGGGGUCGCCGCAAAUGAGACAUGGAAAUAUGGC ....((((.((((((((...((....................))(((((..((((...))))...)))))..))))))))))))..................... (-24.37 = -24.82 + 0.45)
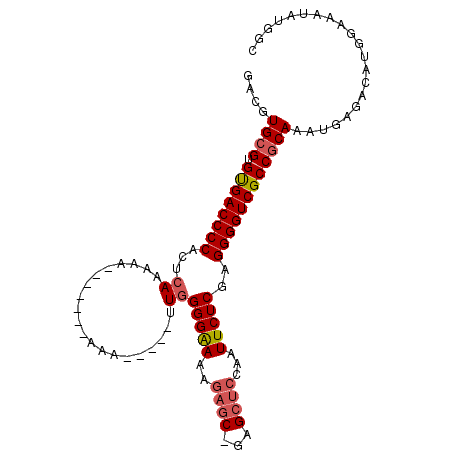
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:24 2006