
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,535,355 – 12,535,453 |
| Length | 98 |
| Max. P | 0.967500 |

| Location | 12,535,355 – 12,535,453 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 85.80 |
| Mean single sequence MFE | -23.72 |
| Consensus MFE | -20.63 |
| Energy contribution | -21.02 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12535355 98 + 23771897 AUGUAUAUAUGCAUAUUCAAAACGCCAGUCGCUGCUAAUA-UGUAGUAGCGGCUUAUAAAAUGUUACAAGUUUUGUAUUAAUUAAAAACUUUGGCUCGU-- (((((....))))).........((((((((((((((...-..)))))))))).((((((((.......))))))))..............))))....-- ( -25.70) >DroPse_CAF1 23359 95 + 1 CUG------CUCGCAUUCAAAACGCCAGUCGCUGCUAAUAUUGUAGUAGCGGCUUAUAAAAUGUUACAAGUUUUGUAUUAAUUAAAAACUUUUGCUUAUGU ...------...((((.(((((....(((((((((((......)))))))))))((((((((.......))))))))............)))))...)))) ( -21.80) >DroEre_CAF1 24856 92 + 1 AUG------UGCAUAUUCAAAACGCCAGUCGCUGCUAAUA-UGUAGUAGCGGCUUAUAAAAUGUUACAAGUUUUGUAUUAAUUAAAAACUUCGGCUCGU-- ...------..............((((((((((((((...-..)))))))))))((((((((.......))))))))...............)))....-- ( -24.10) >DroYak_CAF1 24191 92 + 1 AUG------UGCAUAUUCAAAACGCCAGUCGCUGCUAAUA-UGUAGUAGCGGCUUAUAAAAUGUUACAAGUUUUGUAUUAAUUAAAAACUUCGGCUCGU-- ...------..............((((((((((((((...-..)))))))))))((((((((.......))))))))...............)))....-- ( -24.10) >DroAna_CAF1 21114 86 + 1 CUG------UAC----GCCAGCCGCCGUGUGCUGCUAAUA-UGUAGUAGCGGCUUAUAAAAUGUUACAAGUUUUGUAUUAAUUAAAAACUUUGGCUU---- ...------...----(((((..(((((.((((((.....-.))))))))))).((((((((.......)))))))).............)))))..---- ( -25.10) >DroPer_CAF1 23363 95 + 1 CUA------GUCACAUUCAAAACGCCAGUCGCUGCUAAUAUUGUAGUAGCGGCUUAUAAAAUGUUACAAGUUUUGUAUUAAUUAAAAACUUUUGCCUAUGU ...------...((((.(((((....(((((((((((......)))))))))))((((((((.......))))))))............)))))...)))) ( -21.50) >consensus AUG______UGCAUAUUCAAAACGCCAGUCGCUGCUAAUA_UGUAGUAGCGGCUUAUAAAAUGUUACAAGUUUUGUAUUAAUUAAAAACUUUGGCUCGU__ .......................((((((((((((((......)))))))))))((((((((.......))))))))...............)))...... (-20.63 = -21.02 + 0.39)

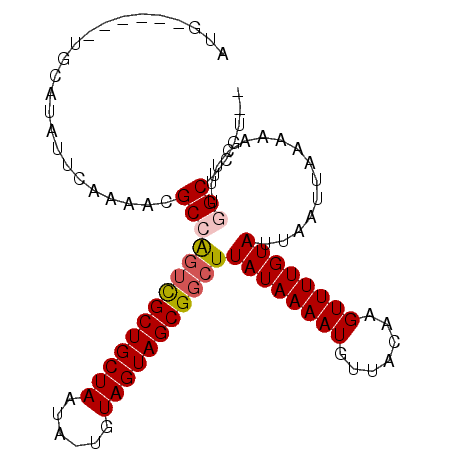

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:21 2006