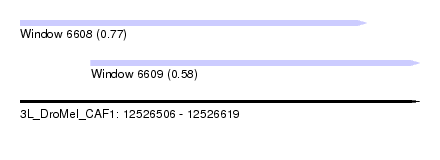
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,526,506 – 12,526,619 |
| Length | 113 |
| Max. P | 0.767775 |

| Location | 12,526,506 – 12,526,604 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.70 |
| Mean single sequence MFE | -13.82 |
| Consensus MFE | -10.50 |
| Energy contribution | -10.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12526506 98 + 23771897 GCCGCAAAAAACA---------------GCAAACUAAAACAGCAAAAAAA--AUAAAGGUAUAAAAACAUUAAACAUUAAACAGCGCUGAAUGUUUGUGGCUAUGUACAAUCGCA ((((((((...((---------------((...((.....))........--......((.(((.....))).))..........))))....)))))))).............. ( -15.20) >DroSec_CAF1 14782 95 + 1 GCCGCAAAACGCC----------------AAAACUAAAACAGCAAAAAAA----AAAUGUAUAAAAACAUUAAACAUUAAACAGCGCUGAAUGUUUGUGGCUAUGUACAAUCGCA ((((((((..(..----------------....).....((((.......----.(((((.(((.....))).))))).......))))....)))))))).............. ( -15.36) >DroSim_CAF1 14574 99 + 1 GCCGCAAAAAGCC----------------AAAACUAAAACAGCAAAAAAAGUAUAAAAGUAUAAAAACAUUAAACAUUAAACAGCGCUGAAUGUUUGUGGCUAUGUACAAUCGCA ((((((((.((..----------------....))....((((.......((((....)))).......(((.....))).....))))....)))))))).............. ( -13.50) >DroEre_CAF1 14697 105 + 1 GCCGCAAAAAAACACAACAAAAAUAUAACCAAACUAAAACAGCAAAAA---------AGUAUAA-AACAUUAAACAUUAAACAGCGCUGAAUGUUUGUGGCUAUGUACAUUCGUA ((((((((.................((.......))...((((.....---------.((....-.)).(((.....))).....))))....)))))))).............. ( -11.20) >consensus GCCGCAAAAAACC________________AAAACUAAAACAGCAAAAAAA____AAAAGUAUAAAAACAUUAAACAUUAAACAGCGCUGAAUGUUUGUGGCUAUGUACAAUCGCA (((((.....................................................((......))...(((((((...........)))))))))))).............. (-10.50 = -10.50 + 0.00)


| Location | 12,526,526 – 12,526,619 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 90.56 |
| Mean single sequence MFE | -14.19 |
| Consensus MFE | -10.81 |
| Energy contribution | -10.62 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12526526 93 + 23771897 AAAACAGCAAAAAAA--AUAAAGGUAUAAAAACAUUAAACAUUAAACAGCGCUGAAUGUUUGUGGCUAUGUACAAUCGCACAGGCAGAAAUGUAC-- ....((((.......--......((......)).(((.....))).....)))).((((((.((.((.(((......))).)).)).))))))..-- ( -12.40) >DroSec_CAF1 14801 93 + 1 AAAACAGCAAAAAAA----AAAUGUAUAAAAACAUUAAACAUUAAACAGCGCUGAAUGUUUGUGGCUAUGUACAAUCGCACAGGCAGAAAUGUACAC ....((((.......----.(((((.(((.....))).))))).......)))).((((((.((.((.(((......))).)).)).)))))).... ( -15.86) >DroSim_CAF1 14593 97 + 1 AAAACAGCAAAAAAAGUAUAAAAGUAUAAAAACAUUAAACAUUAAACAGCGCUGAAUGUUUGUGGCUAUGUACAAUCGCACAAGCAGAAAUGUACAC ...............(((((...(((((.(..(((.(((((((...........))))))))))..).)))))....((....)).....))))).. ( -14.00) >DroEre_CAF1 14732 85 + 1 AAAACAGCAAAAA---------AGUAUAA-AACAUUAAACAUUAAACAGCGCUGAAUGUUUGUGGCUAUGUACAUUCGUACAGGCAGAAAUGUAC-- .............---------.......-.(((((...(((.(((((........))))))))(((.(((((....))))))))...)))))..-- ( -14.50) >consensus AAAACAGCAAAAAAA____AAAAGUAUAAAAACAUUAAACAUUAAACAGCGCUGAAUGUUUGUGGCUAUGUACAAUCGCACAGGCAGAAAUGUAC__ .......................(((((....(((.(((((((...........))))))))))(((.(((.(....).)))))).....))))).. (-10.81 = -10.62 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:18 2006