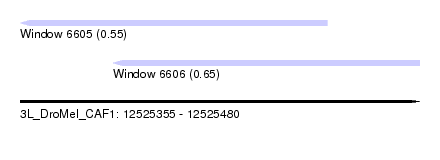
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,525,355 – 12,525,480 |
| Length | 125 |
| Max. P | 0.653992 |

| Location | 12,525,355 – 12,525,451 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 82.79 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -19.31 |
| Energy contribution | -18.73 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
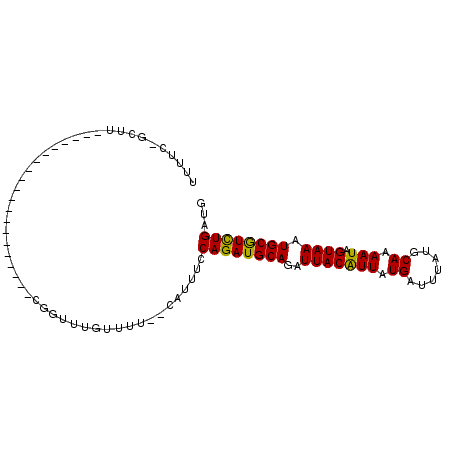
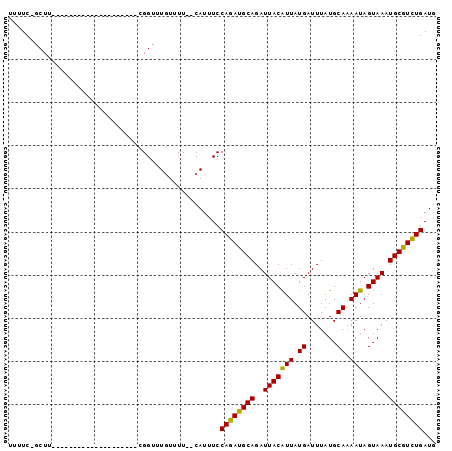
>3L_DroMel_CAF1 12525355 96 - 23771897 -----------CGGUUUGUUUUCAUUUCCAGAUGCAGAUUACAUUAUGAUUUAUGCAAAAUAGUAAAUGCGUCUGAUGGGCAACAUGCGCCAGCUGCCAGACACGA-------A -----------(((((((..........((((((((..(((((((.((.......)).))).)))).)))))))).(((((.....)).))).....))))).)).-------. ( -24.90) >DroVir_CAF1 13619 101 - 1 -----------CGGUUUGUUUUCAUUUCCAGAUGCAGAUUACAUUAUGAUUUAUGCAAAAUAGUAAAUGCGUCUGAUG-GCAACAUGCGCCAGCCGACAGCCGCCAACU-UGUA -----------(((((.(((..(.....((((((((..(((((((.((.......)).))).)))).)))))))).((-((.......)))))..))))))))......-.... ( -25.70) >DroGri_CAF1 13651 95 - 1 -----------CGGUUUGUUUUCAUUUCCAGAUGCAGAUUACAUUAUGAUUUAUGCAAAAUAGUAAAUGCGUCUGAUG-GCAACAUGCGCCAGCCUCCA-------ACUUUGUA -----------.((((.((.........((((((((..(((((((.((.......)).))).)))).))))))))(((-....)))..)).))))....-------........ ( -22.00) >DroWil_CAF1 37349 95 - 1 -----------UGGUUUAUUUUCAUUUCCAGAUGCAGAUUACGUUAUGAUUUAUGCAUAAUAGUAAAUGCAUUUGAUU-GCAACAUGCGCCAACUGGAAGACGUGG-------G -----------..........(((((((((((((((..((((((((((.......)))))).)))).)))))......-((.....)).....))))))...))))-------. ( -20.80) >DroMoj_CAF1 14237 90 - 1 -----------CGGUUUGUUUUCAUUUCCAGAUGCAGAUUACAUUAUGAUUUAUGCAAAAUAGUAAAUGCGUCUGAUG-GCAACAUGCGCCAGCCGCCAUUU------------ -----------(((((.((.........((((((((..(((((((.((.......)).))).)))).))))))))(((-....)))..)).)))))......------------ ( -23.30) >DroAna_CAF1 12252 106 - 1 CGCCUUAGCUUCGGUUUGUUUUCAUUUCCAGAUGCAGAUUACAUUAUGAUUUAUGCAAAAUAGUAAAUGCGUCUGAUG-GCAACAUGCGCCAGCUGCCAGACACGA-------A .((....)).((((((((..........((((((((..(((((((.((.......)).))).)))).)))))))).((-((.......)))).....))))).)))-------. ( -26.90) >consensus ___________CGGUUUGUUUUCAUUUCCAGAUGCAGAUUACAUUAUGAUUUAUGCAAAAUAGUAAAUGCGUCUGAUG_GCAACAUGCGCCAGCCGCCAGAC_CGA_______A ...........(((((.((.........((((((((..(((((((.((.......)).))).)))).))))))))....((.....)))).))))).................. (-19.31 = -18.73 + -0.58)
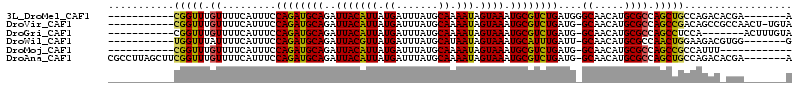
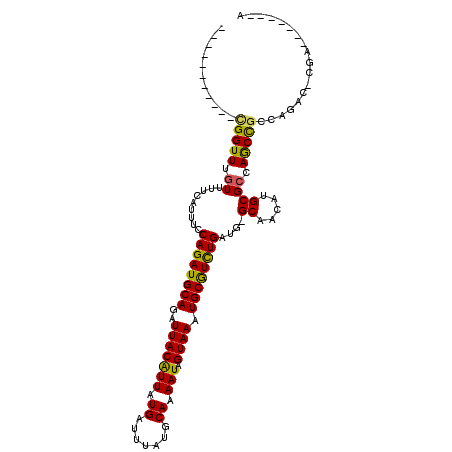
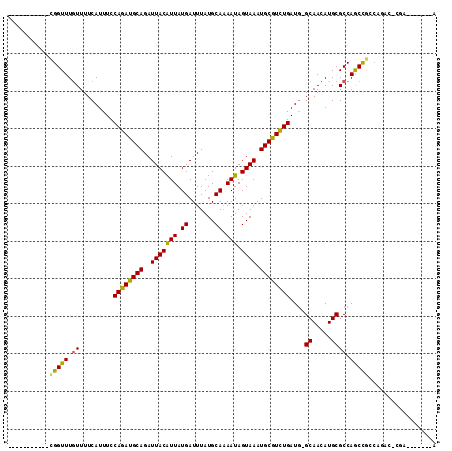
| Location | 12,525,384 – 12,525,480 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 87.33 |
| Mean single sequence MFE | -15.87 |
| Consensus MFE | -14.28 |
| Energy contribution | -13.87 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12525384 96 - 23771897 UUUUC-GCUUGAGUUCGCCUCGCAUCGCAUCGGUUUGUUUU--CAUUUCCAGAUGCAGAUUACAUUAUGAUUUAUGCAAAAUAGUAAAUGCGUCUGAUG .....-((..(((.....)))))....((((((..((....--))...))(((((((..(((((((.((.......)).))).)))).))))))))))) ( -18.40) >DroVir_CAF1 13653 77 - 1 UUUUCAGCUU--------------------CGGUUUGUUUU--CAUUUCCAGAUGCAGAUUACAUUAUGAUUUAUGCAAAAUAGUAAAUGCGUCUGAUG .....(((..--------------------..)))......--......((((((((..(((((((.((.......)).))).)))).))))))))... ( -16.50) >DroPse_CAF1 12912 78 - 1 UUUUC-GCUU--------------------UGGUUUGUUUUCUCAUUUCCAGAUGCAGAUUACAUUAUGAUUUAUGCAAAAUAGUAAAUGCGUCUGAUG .....-....--------------------...................((((((((..(((((((.((.......)).))).)))).))))))))... ( -14.30) >DroWil_CAF1 37377 76 - 1 UUUUC-GCUU--------------------UGGUUUAUUUU--CAUUUCCAGAUGCAGAUUACGUUAUGAUUUAUGCAUAAUAGUAAAUGCAUUUGAUU .....-....--------------------...........--......((((((((..((((((((((.......)))))).)))).))))))))... ( -15.80) >DroMoj_CAF1 14260 76 - 1 UUUUC-GCUU--------------------CGGUUUGUUUU--CAUUUCCAGAUGCAGAUUACAUUAUGAUUUAUGCAAAAUAGUAAAUGCGUCUGAUG .....-((..--------------------..)).......--......((((((((..(((((((.((.......)).))).)))).))))))))... ( -15.90) >DroPer_CAF1 12737 78 - 1 UUUUC-GCUU--------------------UGGUUUGUUUUCUCAUUUCCAGAUGCAGAUUACAUUAUGAUUUAUGCAAAAUAGUAAAUGCGUCUGAUG .....-....--------------------...................((((((((..(((((((.((.......)).))).)))).))))))))... ( -14.30) >consensus UUUUC_GCUU____________________CGGUUUGUUUU__CAUUUCCAGAUGCAGAUUACAUUAUGAUUUAUGCAAAAUAGUAAAUGCGUCUGAUG .................................................((((((((..(((((((.((.......)).))).)))).))))))))... (-14.28 = -13.87 + -0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:16 2006