
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,484,061 – 12,484,214 |
| Length | 153 |
| Max. P | 0.965714 |

| Location | 12,484,061 – 12,484,181 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -28.76 |
| Consensus MFE | -21.64 |
| Energy contribution | -21.12 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12484061 120 - 23771897 UAUAUUUUAGCUCAAAGUUGAGGACCUAUCCGCCAAGAAGAAAAAGACAUUGGCGGACAAACUUUUCGGUUCUAGCCACACAAACAAGGAGAACGUGUCACCCAAUGACGUGGAAUCUUC ....(((((((.....))))))).....((((((((.............))))))))..........(((....)))..........(((((.(((((((.....)))))))...))))) ( -26.32) >DroVir_CAF1 21215 119 - 1 -UUUUUUCAGCUUAAAGUCGAGGAUUUGUCAGCCAAGAAGAAGAAAACUUUGGCCGACAAAAUCUUUGGAUCCAACAGCAAUACUAAGGAGAACAUAUCGCCCAACGAUGUAGAAUCAUC -........(((.....(((((((((((((.((((((...........)))))).))).)))))))))).......))).........((....((((((.....))))))....))... ( -27.60) >DroEre_CAF1 10475 118 - 1 --UAUUUUAGCUUAAGGUAGAGGACCUGUCUUCCAAGAAGAAAAAGACAUUGGCGGACAAGCUCUUCGGUUCCAGCCAUACAAACAAGGAGAACAUGUCACCCAAUGACGUGGAAUCUUC --..............(((((((((.(((((.((((.............)))).))))).).)))))(((....))).)))......(((((.(((((((.....)))))))...))))) ( -30.32) >DroYak_CAF1 9542 118 - 1 --UCUAUUAGCUUAAAGUAGAGGACUUGUCUUCCAAGAAGAAAAAGACUUUGGCGGACAAGCUCUUCGGUUCCAGCCACACAAACAAGGAGAACGUGUCACCCAAUGACGUGGAAUCUUC --.................((((((((((((.(((((...........))))).))))))).)))))(((((((.((..........)).......((((.....)))).)))))))... ( -30.80) >DroMoj_CAF1 11579 119 - 1 -AAAUUGUAGCUUAAGGUGGAGGAUUUGUCUGCGAAGAAGAAGAAAACACUUGCAGACAAAAUCUUUGGAUCCAACAGCAAUACCAAGGAGAACAUAUCGCCUAAUGAUGUGGAGUCAUC -.............((((((....(((((((((((...............))))))))))).(((((((..............))))))).......)))))).(((((.....))))). ( -28.60) >DroAna_CAF1 8509 120 - 1 UGAAUUUCAGCUCAAAGUAGAAGACUUGUCUUCAAAGAAAAAGAAGACCCUAGCGGACAAGCUCUUCGGCUCCAGUCACACCAAUAAGGAGAACGUGUCACCCAAUGACGUGGAGUCCUC ...................((((((((((((((.........))))))((....))..))).)))))(((((((((((..((.....)).((.....))......)))).)))))))... ( -28.90) >consensus __UAUUUUAGCUUAAAGUAGAGGACUUGUCUGCCAAGAAGAAAAAGACAUUGGCGGACAAACUCUUCGGUUCCAGCCACACAAACAAGGAGAACAUGUCACCCAAUGACGUGGAAUCUUC ...................((((((((((((.((((.............)))).))))))).)))))(((((((.............(.....).(((((.....))))))))))))... (-21.64 = -21.12 + -0.52)

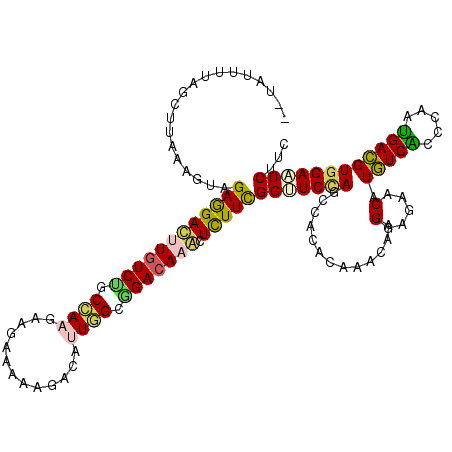

| Location | 12,484,101 – 12,484,214 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 72.37 |
| Mean single sequence MFE | -23.06 |
| Consensus MFE | -17.75 |
| Energy contribution | -18.03 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12484101 113 + 23771897 UGUGGCUAGAACCGAAAAGUUUGUCCGCCAAUGUCUUUUUCUUCUUGGCGGAUAGGUCCUCAACUUUGAGCUAAAAUAUAAAUACAAAAAUGUUUUUAAUCUCAUUAACUAUU ..(((((...........(..((((((((((.............))))))))))..)...........))))).....((((.(((....))).))))............... ( -21.47) >DroVir_CAF1 21255 100 + 1 UGCUGUUGGAUCCAAAGAUUUUGUCGGCCAAAGUUUUCUUCUUCUUGGCUGACAAAUCCUCGACUUUAAGCUGAAAAAA--------AUAUGCAUUGGAG---AACAAGCA-- .((((((...(((((((..((((((((((((((.......))..))))))))))))..)).........((........--------....)).))))).---))).))).-- ( -27.80) >DroSec_CAF1 9576 113 + 1 UGUGGCUAGAACCGAAAAGCUUGUCCGCCAAUGUCUUUUUCUUCUUGGCGGACAGGUCCUCAACUUUGAGCUAAAAUAUAAAUUUAAAAAUGUAUGUAAUCUCAUUAAAUAUU ..(((((...........(((((((((((((.............)))))))))))))...........)))))..(((((.(((....))).)))))................ ( -26.57) >DroEre_CAF1 10515 105 + 1 UAUGGCUGGAACCGAAGAGCUUGUCCGCCAAUGUCUUUUUCUUCUUGGAAGACAGGUCCUCUACCUUAAGCUAAAAUA--------GACAUGUAUUCAGGUUCAUUAAAUAUU ........((((((((..((.((((......((((((((.......))))))))(((.....))).............--------)))).)).))).))))).......... ( -22.70) >DroYak_CAF1 9582 105 + 1 UGUGGCUGGAACCGAAGAGCUUGUCCGCCAAAGUCUUUUUCUUCUUGGAAGACAAGUCCUCUACUUUAAGCUAAUAGA--------AAAAUGUAUUUAGUUUCAUAAAAUAUU ..(((((.(((....((..((((((..((((((.......))..))))..))))))..))....))).))))).....--------..(((((.((((......))))))))) ( -19.30) >DroMoj_CAF1 11619 101 + 1 UGCUGUUGGAUCCAAAGAUUUUGUCUGCAAGUGUUUUCUUCUUCUUCGCAGACAAAUCCUCCACCUUAAGCUACAAUUU--------UGGUGUAUUAGUU---AUCAAUUAU- .(((...((......((..((((((((((((............))).)))))))))..))...))...))).......(--------(((((.......)---)))))....- ( -20.50) >consensus UGUGGCUGGAACCGAAGAGCUUGUCCGCCAAUGUCUUUUUCUUCUUGGCAGACAAGUCCUCAACUUUAAGCUAAAAUAU_______AAAAUGUAUUUAAU_UCAUUAAAUAUU ..(((((........((.(((((((((((((.............))))))))))))).))........)))))........................................ (-17.75 = -18.03 + 0.28)
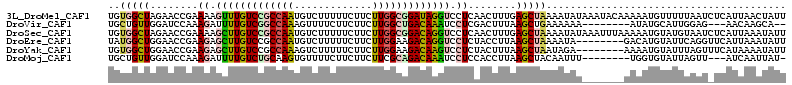

| Location | 12,484,101 – 12,484,214 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 72.37 |
| Mean single sequence MFE | -23.14 |
| Consensus MFE | -15.17 |
| Energy contribution | -14.84 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12484101 113 - 23771897 AAUAGUUAAUGAGAUUAAAAACAUUUUUGUAUUUAUAUUUUAGCUCAAAGUUGAGGACCUAUCCGCCAAGAAGAAAAAGACAUUGGCGGACAAACUUUUCGGUUCUAGCCACA ....(((..((((((((((.(((....))).)))).)))))).((((....)))))))...((((((((.............))))))))..........(((....)))... ( -22.72) >DroVir_CAF1 21255 100 - 1 --UGCUUGUU---CUCCAAUGCAUAU--------UUUUUUCAGCUUAAAGUCGAGGAUUUGUCAGCCAAGAAGAAGAAAACUUUGGCCGACAAAAUCUUUGGAUCCAACAGCA --((((((..---...))).)))...--------........(((.....(((((((((((((.((((((...........)))))).))).)))))))))).......))). ( -24.60) >DroSec_CAF1 9576 113 - 1 AAUAUUUAAUGAGAUUACAUACAUUUUUAAAUUUAUAUUUUAGCUCAAAGUUGAGGACCUGUCCGCCAAGAAGAAAAAGACAUUGGCGGACAAGCUUUUCGGUUCUAGCCACA .........((((.....(((.(((....))).))).......))))..(((.((((((((((((((((.............))))))))))........))))))))).... ( -24.52) >DroEre_CAF1 10515 105 - 1 AAUAUUUAAUGAACCUGAAUACAUGUC--------UAUUUUAGCUUAAGGUAGAGGACCUGUCUUCCAAGAAGAAAAAGACAUUGGCGGACAAGCUCUUCGGUUCCAGCCAUA ..........(((((.(((...(((((--------(.((((..((((((..((.....))..)))..)))..)))).)))))).(((......))).))))))))........ ( -22.10) >DroYak_CAF1 9582 105 - 1 AAUAUUUUAUGAAACUAAAUACAUUUU--------UCUAUUAGCUUAAAGUAGAGGACUUGUCUUCCAAGAAGAAAAAGACUUUGGCGGACAAGCUCUUCGGUUCCAGCCACA ..(((((((.....((((.........--------....))))..)))))))((((((((((((.(((((...........))))).))))))).)))))(((....)))... ( -23.22) >DroMoj_CAF1 11619 101 - 1 -AUAAUUGAU---AACUAAUACACCA--------AAAUUGUAGCUUAAGGUGGAGGAUUUGUCUGCGAAGAAGAAGAAAACACUUGCAGACAAAAUCUUUGGAUCCAACAGCA -.........---......((((...--------....))))(((...(((.((((((((((((((((...............)))))))).)))))))).)..))...))). ( -21.66) >consensus AAUAUUUAAUGA_AUUAAAUACAUUUU_______AUAUUUUAGCUUAAAGUAGAGGACCUGUCUGCCAAGAAGAAAAAGACAUUGGCGGACAAACUCUUCGGUUCCAGCCACA ....................................................((((((.((((((((((.............)))))))))).).)))))............. (-15.17 = -14.84 + -0.33)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:01 2006