
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,479,776 – 12,479,892 |
| Length | 116 |
| Max. P | 0.637073 |

| Location | 12,479,776 – 12,479,892 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.24 |
| Mean single sequence MFE | -36.93 |
| Consensus MFE | -20.65 |
| Energy contribution | -21.10 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.637073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
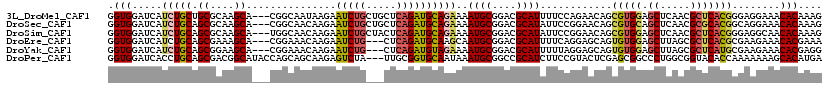
>3L_DroMel_CAF1 12479776 116 - 23771897 GGUGGAUCAUCUGCUGCGCAAGCA---CGGCAAUAAGAAUCUGCUGCUCAGAUGCAGAAAAUGCGGACGCAUUUUCCAGAACAGCGUGGAGCUCAACGCUCACGGGAGGAAACACAAAG .(((..((.((((((((....)).---.)))........((((.(((......)))((((((((....))))))))))))....((((.(((.....)))))))))).))..))).... ( -37.20) >DroSec_CAF1 5247 116 - 1 GGUGGAUCAUCUGCAGCGCAAGCA---CGGCAACAAGAAUCUGCUGCUCAGAUGCAGAAAAUGCGGACGCAUAUUCCGGAACAGCGUGCAGCUCAACGCGCACGGCAGGAAACACAAAG .(((..((.((((((((....)).---(((((.........)))))......))))))...((((((.......))).......(((((.((.....)))))))))).))..))).... ( -37.60) >DroSim_CAF1 4970 116 - 1 GGUGGAUCAUCUGCAGCGCAAGCA---UGGCAACAAGAAUCUGCUACUCAGAUGCAGAAAAUGCGGACGCAUAUUCCGGAACAGCGUGGAGCUCAACGCUCACGGGAGGCAACACAAAG .(((.....((((((((....)).---((....))....(((((.........)))))...)))))).((...(((((....(((((........)))))..))))).))..))).... ( -39.60) >DroEre_CAF1 6107 113 - 1 GGUGGAUCAUCUGCAGCGAAAGCA---CGGAAACAAGAAUCUG---CUCAGAUGCAAGCAAUGCGGACGCAUUUUCAGGAGCAGUGUGGAGCUUAGCGCUCACGCGAAGAAACACGAAA .(((..((.(((...((....)).---.(....).)))..(((---(((...((.((..(((((....))))))))).))))))((((((((.....)))).))))..))..))).... ( -35.20) >DroYak_CAF1 5184 113 - 1 GGUGGAUCAUCUGCAGCGGAAGCA---CGGAAACAAGAAUCUG---CUCAGAUGUAGAAAAUGCGGACGCAUUUUUAGGAGCAGUGUGGAGCUUAGCGCUCAUGCGAAGAAACACGAGG .(((..(((((((.((((((..(.---.(....)..)..))))---)))))))...((((((((....))))))))....(((.....((((.....)))).)))...))..))).... ( -37.00) >DroPer_CAF1 5010 116 - 1 GGUGGAUCACCUGCAGCGACGGCAUACCAGCAGCAAGAGUCUA---UUGCGGUGCAAUAAAUGCGGCCGCAUCUUCCGUACUCGAGCGGCCCUGGCGGUACACCAAAAAAAGCACAUGA ((((.(((.((((((.(....((......)).((((.......---))))).))))........((((((.((..........))))))))..)).))).))))............... ( -35.00) >consensus GGUGGAUCAUCUGCAGCGCAAGCA___CGGCAACAAGAAUCUG___CUCAGAUGCAGAAAAUGCGGACGCAUUUUCCGGAACAGCGUGGAGCUCAACGCUCACGGGAAGAAACACAAAG .(((.....(((((.((....))...............(((((.....))))))))))..((((....))))............(((((.((.....)))))))........))).... (-20.65 = -21.10 + 0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:58 2006