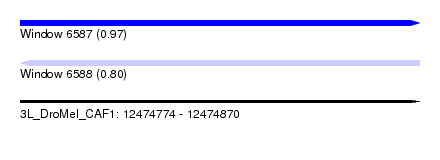
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,474,774 – 12,474,870 |
| Length | 96 |
| Max. P | 0.967851 |

| Location | 12,474,774 – 12,474,870 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 70.47 |
| Mean single sequence MFE | -19.32 |
| Consensus MFE | -9.92 |
| Energy contribution | -11.67 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12474774 96 + 23771897 UUACAUGUGCUGGUAACUAUAUAUA----------ACCCAUCUGUACUAUAUCCUACCUUUGUGACCAGAAACACCUGUUCGCGUUUCAGUUUCUGGUCGCCAAUA .((((.(((..(((...........----------)))))).))))...............((((((((((((................))))))))))))..... ( -21.89) >DroVir_CAF1 11868 95 + 1 UGAUAG--------AUGUGUGGAAAAUGUCUCCAAUUCCAUUU---CAACAUCUUACUUUUGUCACUAGGAAAAUAUCUUCACGCUUGACCACACCCUCAGCAAUC (((.((--------((((.((((.......)))).((((....---..(((.........))).....)))).))))))))).(((.((........))))).... ( -13.60) >DroSec_CAF1 198 96 + 1 AUACAUGGGCUGGUAACUAUAUAUA----------CACCAUCUAUACUAUAUCCUACCUUUGUGACCAGAAACACCUGUUCGCGUUUUAGUUUCUGGUCGCCAAUA .......(((.((((..((((((((----------.......)))).))))...)))).....((((((((((................))))))))))))).... ( -23.29) >DroSim_CAF1 196 96 + 1 AUACAUGGGCUGGUAACUAUAUAUA----------CACCAUCUACACUAUAUCCUACCUUUGUGACCAGAAACACCUGUUCGCGUUUCAGUUUCUGGUCGCCAAUA .......(((.((((...(((((..----------............)))))..)))).....((((((((((................))))))))))))).... ( -22.53) >DroEre_CAF1 196 85 + 1 AUACAUGUUCUGGUAACAA---------------------UAUAUACUAUAUCCUACCUUUGUGACCAGAAACACCUGUUCGCGUCUUAGUUUCUGGUCGCCUAUA ...........((((...(---------------------((((...)))))..))))...((((((((((((................))))))))))))..... ( -20.59) >DroYak_CAF1 196 96 + 1 AUACAUGUUCUGGUAACUAGAUAUC----------CACUUUAUAUACUAUAUCCUACCUUGGUGACCAGAAACACCUGUUCGCGUUUUAUAAUCUUGUCGCCUAUA ...........((..((.((((...----------..................(......)((((.(((......))).))))........)))).))..)).... ( -14.00) >consensus AUACAUGUGCUGGUAACUAUAUAUA__________CACCAUCUAUACUAUAUCCUACCUUUGUGACCAGAAACACCUGUUCGCGUUUUAGUUUCUGGUCGCCAAUA .............................................................((((((((((((................))))))))))))..... ( -9.92 = -11.67 + 1.75)

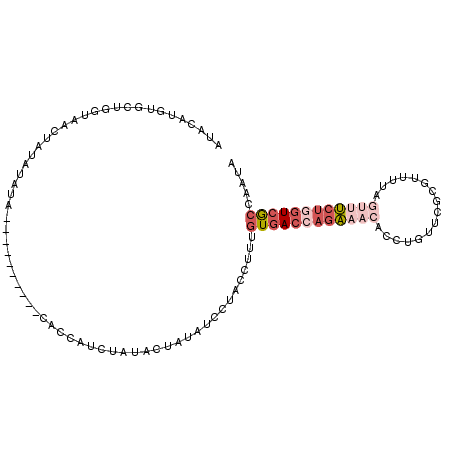
| Location | 12,474,774 – 12,474,870 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 70.47 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -9.62 |
| Energy contribution | -9.65 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12474774 96 - 23771897 UAUUGGCGACCAGAAACUGAAACGCGAACAGGUGUUUCUGGUCACAAAGGUAGGAUAUAGUACAGAUGGGU----------UAUAUAUAGUUACCAGCACAUGUAA ...((((((((((((((................)))))))))).....(((((.((((((..(....)..)----------)))))....))))).)).))..... ( -25.89) >DroVir_CAF1 11868 95 - 1 GAUUGCUGAGGGUGUGGUCAAGCGUGAAGAUAUUUUCCUAGUGACAAAAGUAAGAUGUUG---AAAUGGAAUUGGAGACAUUUUCCACACAU--------CUAUCA .......(((((((((((((...(.((((....)))))...))))...............---...(((((.((....))..))))))))))--------)).)). ( -22.60) >DroSec_CAF1 198 96 - 1 UAUUGGCGACCAGAAACUAAAACGCGAACAGGUGUUUCUGGUCACAAAGGUAGGAUAUAGUAUAGAUGGUG----------UAUAUAUAGUUACCAGCCCAUGUAU (((.(((((((((((((................)))))))))).....(((((.(((((.((((.....))----------)))))))..))))).))).)))... ( -27.99) >DroSim_CAF1 196 96 - 1 UAUUGGCGACCAGAAACUGAAACGCGAACAGGUGUUUCUGGUCACAAAGGUAGGAUAUAGUGUAGAUGGUG----------UAUAUAUAGUUACCAGCCCAUGUAU (((.(((((((((((((................)))))))))).....(((((.(((((............----------..)))))..))))).))).)))... ( -26.63) >DroEre_CAF1 196 85 - 1 UAUAGGCGACCAGAAACUAAGACGCGAACAGGUGUUUCUGGUCACAAAGGUAGGAUAUAGUAUAUA---------------------UUGUUACCAGAACAUGUAU .....((((((((((((................)))))))))).....(((((((((((...))))---------------------)).))))).......)).. ( -21.59) >DroYak_CAF1 196 96 - 1 UAUAGGCGACAAGAUUAUAAAACGCGAACAGGUGUUUCUGGUCACCAAGGUAGGAUAUAGUAUAUAAAGUG----------GAUAUCUAGUUACCAGAACAUGUAU ....((.(((.(((......(((((......)))))))).))).))..((((((((((..(((.....)))----------.))))))...))))........... ( -18.90) >consensus UAUUGGCGACCAGAAACUAAAACGCGAACAGGUGUUUCUGGUCACAAAGGUAGGAUAUAGUAUAGAUGGUG__________UAUAUAUAGUUACCAGCACAUGUAU .......(((((((......(((((......))))))))))))............................................................... ( -9.62 = -9.65 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:57 2006