| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,464,843 – 12,464,939 |
| Length | 96 |
| Max. P | 0.986712 |

| Location | 12,464,843 – 12,464,939 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 73.43 |
| Mean single sequence MFE | -25.83 |
| Consensus MFE | -17.27 |
| Energy contribution | -18.97 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12464843 96 - 23771897 UUCUUUGGAUUUAGGUGUGAGCGUUUGUUUGUGCCCGACACUCGUGCUCACAAGUGAAGUCAACGAAAUACAAUUU----------ACUAGUUAAAUAAUAACUAA------- (((.(((..((((..((((((((..((..(((.....)))..))))))))))..))))..))).))).........----------..((((((.....)))))).------- ( -22.80) >DroSec_CAF1 19267 93 - 1 UUCUUUGGCUUUAGUUGUGAGCGUUUGUUUGUGCCCGACACUCGUGCUCACAAGUGGAGUCAAUGAAAUAAAAUUC----------ACUAAUGAAAU---AAAUAA------- (((.(((((((((.(((((((((..((..(((.....)))..))))))))))).))))))))).)))......(((----------(....))))..---......------- ( -28.10) >DroSim_CAF1 19501 93 - 1 UUCUUUGGCUUUAGGUGUGAGCGUUUGUUUGUGCCCGACACUCGUGCUCACAAGUGGAGUCAAUGAAAUACAAUUU----------ACUAAUGAAAU---AAAUAA------- (((.(((((((((..((((((((..((..(((.....)))..))))))))))..))))))))).))).........----------...........---......------- ( -25.50) >DroEre_CAF1 18908 109 - 1 UUCUUUGGCUUUAGGUGUGAGCGUUUGUUUGUGU-CGACACUCGUGCUCACAAGUGGGGUCAAAGAAAUACCAUUUAUACUUAGUUACCAGUUAAAU---AACGAAUUACCUU (((((((((((((..((((((((..((...(((.-...))).))))))))))..))))))))))))).....(((((.(((........))))))))---............. ( -30.10) >DroYak_CAF1 19330 109 - 1 UUCUUUGGCUUUAGGUGUGAGCGCUUGUUUGUGU-CGACACUCGUGCUCACAAGUGGGGUCAAUGAAAUACAAUUUGCACUUCGUUGCCAGUCAAAU---AAUUAAUUACCUU (((.(((((((((..(((((((((......(((.-...)))..)))))))))..))))))))).))).....(((((.(((........))))))))---............. ( -30.90) >DroAna_CAF1 17580 89 - 1 UUCUUGGGCUUGAAGUGUG----UUUGUUUCUUAUAGACACUCUUGACAACAAUUGGCGGCAAAACAAUAGGGCUU----------UAUAGGAAAUU---AAAUAU------- (((((((((((.(((.(((----(((((.....)))))))).))).......((((.........)))).))))).----------..))))))...---......------- ( -17.60) >consensus UUCUUUGGCUUUAGGUGUGAGCGUUUGUUUGUGCCCGACACUCGUGCUCACAAGUGGAGUCAAUGAAAUACAAUUU__________ACUAGUGAAAU___AAAUAA_______ (((.(((((((((..((((((((..((((.......))))....))))))))..))))))))).))).............................................. (-17.27 = -18.97 + 1.69)

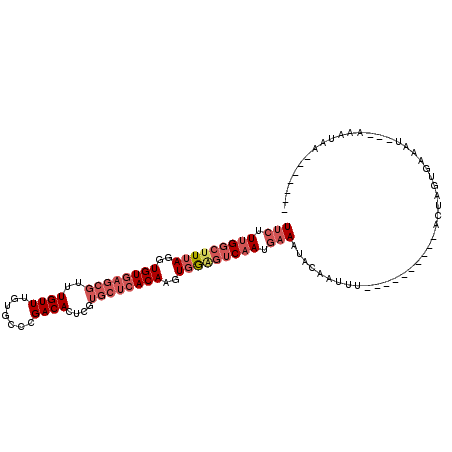

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:52 2006