
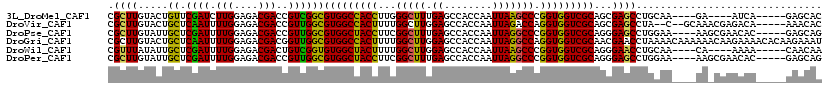
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,439,022 – 12,439,128 |
| Length | 106 |
| Max. P | 0.625854 |

| Location | 12,439,022 – 12,439,128 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.40 |
| Mean single sequence MFE | -37.67 |
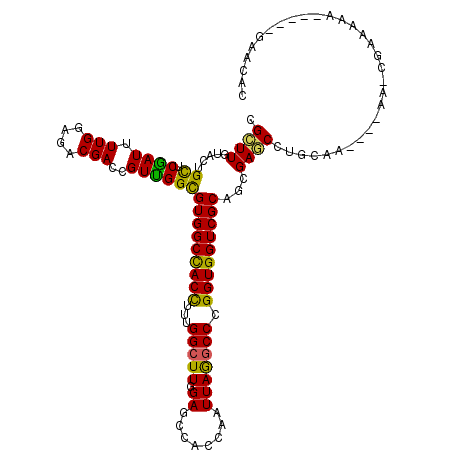
| Consensus MFE | -29.68 |
| Energy contribution | -28.35 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12439022 106 + 23771897 CGCUUGUACUGUUCGAUCUUGGAGACGACCGUCGGCGUGGCCACCUUGGGCUUUGAGCCACCAAUUAAGCCCGGUGGUCGCAGCGAGCCUGCAA----GA----AUCA-----GAGCAC ..((((((..(((((((.(((....)))..))).(((((((((((..((((((.............))))))))))))))).)))))).)))))----).----....-----...... ( -40.82) >DroVir_CAF1 41790 110 + 1 CGCUUGUACUGCUCAAUUUUGGAGACGACCGUUGGCGUGGCCACUUUUGGCUUGGAGCCACCAAUUAGACCAGGUGGUCGCAGCGAGCCUA--C--GCAAACGAGACA-----AAACAC .((((((...((.((((.(((....)))..))))))(((((((((..((((((((.....)))...)).)))))))))))).))))))...--.--............-----...... ( -35.10) >DroPse_CAF1 42114 110 + 1 CGCUUGUAUUGCUCGAUUUUGGAGACGACCGUUGGCGUGGCUACCUUCGGCUUUGAGCCACCAAUUAGGCCCGGUGGUCGCAGGGAGCCUGGAA----AAGCGAACAC-----GAGCAG .........((((((...(((....)))..(((.((((((((..(....).....))))))...((((((((.((....))..)).))))))..----..)).))).)-----))))). ( -40.50) >DroGri_CAF1 45111 119 + 1 CGCUUGUACUGCUCAAUUUUGGAGACGACGGUUGGCGUGGCCACUUUUGGCUUGGAGCCACCAAUUAGGCCAGGUGGUCGCAACGAACCUAAAACAAAAAACAAGAAAACACAAGAAAU ..(((((...((.((((((((....))).)))))))(((((((((..((((((((.....)))....))))))))))))))...................))))).............. ( -36.30) >DroWil_CAF1 66651 106 + 1 CGUUUAUAUUGCUCGAUUUUGGAGACGACUGUCGGUGUGGCUACUUUUGGCUUGGAGCCACCAAUUAAGCCCGGUGGUCGCAGGGAACCUGCAA----CA----AAAA-----CAACAA .((((.(.(((((((((.(((....)))..)))))((((((((((...(((((((.....)))....)))).))))))))))........))))----.)----.)))-----)..... ( -32.80) >DroPer_CAF1 42566 110 + 1 CGCUUGUAUUGCUCGAUUUUGGAGACGACCGUUGGCGUGGCUACCUUCGGCUUUGAGCCACCAAUUAGGCCCGGUGGUCGCAGGGAGCCUGGAA----AAGCGAACAC-----GAGCAG .........((((((...(((....)))..(((.((((((((..(....).....))))))...((((((((.((....))..)).))))))..----..)).))).)-----))))). ( -40.50) >consensus CGCUUGUACUGCUCGAUUUUGGAGACGACCGUUGGCGUGGCCACCUUUGGCUUGGAGCCACCAAUUAGGCCCGGUGGUCGCAGCGAGCCUGCAA____AA_CGAAAAA_____GAACAC .((((.....((.((((.(((....)))..))))))(((((((((...(((((.((........))))))).)))))))))...))))............................... (-29.68 = -28.35 + -1.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:46 2006