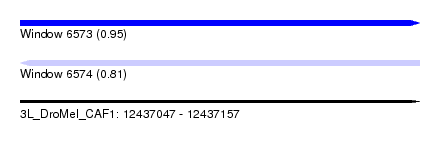
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,437,047 – 12,437,157 |
| Length | 110 |
| Max. P | 0.953394 |

| Location | 12,437,047 – 12,437,157 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 93.94 |
| Mean single sequence MFE | -32.17 |
| Consensus MFE | -29.41 |
| Energy contribution | -28.97 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12437047 110 + 23771897 GUUGCAAUUUCAAAUGCAAUUGCAAUUGCAUCAGUAGUUGCUGUUGCUUUUGCUGCUUUUGUUUUUGUUGCCACUGUCAAUCACAGAGGCGCGCACAACUCAGGUUUUCA ((((...........(((((.((((((((....)))))))).)))))...((((((((((((..(((..........)))..))))))))).)))))))........... ( -32.40) >DroSec_CAF1 39912 104 + 1 GCUGCAAUUUCAAAUGCAAUUGCAAUUGCAUCAGUAGUUGUUGUUGC------UGCUUUUGUUUUUGUUGCCACUGUCAAUCACAGAGGCGCGCACAACUCAGGUUUUCA ((.((((...((((.(((((.((((((((....)))))))).)))))------....))))...)))).(((.((((.....)))).))))).................. ( -29.20) >DroSim_CAF1 40654 109 + 1 GCUGCA-UUUCAAAUGCAAUUGCAAUUGCAUCAGUAGUUGCUGUUGCUGCUGCUGCUUUUGUUUUUGUUGCCACUGUCAAUCACAGAGGCGCGCACAACUCAGGUUUUCA ((.((.-........((((..((((..(((.(((((((.......))))))).)))..))))..)))).(((.((((.....)))).)))))))................ ( -34.90) >consensus GCUGCAAUUUCAAAUGCAAUUGCAAUUGCAUCAGUAGUUGCUGUUGCU__UGCUGCUUUUGUUUUUGUUGCCACUGUCAAUCACAGAGGCGCGCACAACUCAGGUUUUCA ((.((..........(((((.((((((((....)))))))).))))).......((((((((..(((..........)))..))))))))))))................ (-29.41 = -28.97 + -0.44)

| Location | 12,437,047 – 12,437,157 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 93.94 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -25.32 |
| Energy contribution | -25.43 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12437047 110 - 23771897 UGAAAACCUGAGUUGUGCGCGCCUCUGUGAUUGACAGUGGCAACAAAAACAAAAGCAGCAAAAGCAACAGCAACUACUGAUGCAAUUGCAAUUGCAUUUGAAAUUGCAAC ...........(((((....(((.((((.....)))).)))........((((.(((((((..(((.(((......))).)))..))))...))).)))).....))))) ( -29.10) >DroSec_CAF1 39912 104 - 1 UGAAAACCUGAGUUGUGCGCGCCUCUGUGAUUGACAGUGGCAACAAAAACAAAAGCA------GCAACAACAACUACUGAUGCAAUUGCAAUUGCAUUUGAAAUUGCAGC .......(((.((((((((((((.((((.....)))).))).............)).------)).))))).....(.((((((((....)))))))).)......))). ( -25.11) >DroSim_CAF1 40654 109 - 1 UGAAAACCUGAGUUGUGCGCGCCUCUGUGAUUGACAGUGGCAACAAAAACAAAAGCAGCAGCAGCAACAGCAACUACUGAUGCAAUUGCAAUUGCAUUUGAAA-UGCAGC .......(((.(((((..(((((.((((.....)))).))).............((....)).)).))))).....(.((((((((....)))))))).)...-..))). ( -29.10) >consensus UGAAAACCUGAGUUGUGCGCGCCUCUGUGAUUGACAGUGGCAACAAAAACAAAAGCAGCA__AGCAACAGCAACUACUGAUGCAAUUGCAAUUGCAUUUGAAAUUGCAGC ..........((((((....(((.((((.....)))).))).............((.......))....)))))).....(((((((.(((......))).))))))).. (-25.32 = -25.43 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:43 2006