
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,434,611 – 12,434,709 |
| Length | 98 |
| Max. P | 0.562810 |

| Location | 12,434,611 – 12,434,709 |
|---|---|
| Length | 98 |
| Sequences | 6 |
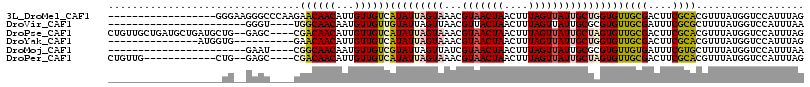
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.69 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -14.42 |
| Energy contribution | -14.78 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12434611 98 + 23771897 ------------------GGGAAGGGCCCAAGAACAACAUUGUUGUCAUAUUAGUAAACGUAACUAACUUUAGUUAUUGCUGGUGUUGCGACUUCGCACGUUUAUGGUCCAUUUAG ------------------.....(((((...((((......(((((.(((((((((...(((((((....)))))))))))))))).))))).......))))..)))))...... ( -27.62) >DroVir_CAF1 36949 89 + 1 -----------------------GGGU----UGGCAACAAUGUUGUUGUAUUAGUUAACGUUACUAACUUUAGUUAUUGCGCGUGUUGCGAUUUCGCGCUUUUAUGGUCCAUUUAA -----------------------(((.----.(((((((....)))))).(((((.......)))))...........(((((...........))))).....)..)))...... ( -17.80) >DroPse_CAF1 37414 110 + 1 CUGUUGCUGAUGCUGAUGCUG--GAGC----CGACAACAUUGUUGUCAUAUUAGUAAACGUAACUAACUUUAGUUAUUGCUAGUGUUGCGACUUCGCACGUUUAUGGUCCAUUUAG ......((((.((....))((--((.(----(((((((...))))))......((((((((....(((.(((((....))))).)))(((....))))))))))))))))).)))) ( -30.00) >DroYak_CAF1 39699 91 + 1 ---------------AUGGUG----------GAACAACAUUGUUGUCAUAUUAGUAAACGUAACUAACUUUAGUUAUUGCUGGUGUUGCGACUUCGCACGUUUAUGGUCCAUUUAG ---------------..((((----------(((((((...))))).......((((((((....(((.(((((....))))).)))(((....)))))))))))..))))))... ( -22.40) >DroMoj_CAF1 32921 89 + 1 -----------------------GAAU----CGGCAACAAUGUUGUCGUAUUAGUUAUCGUAACUAACUUUAGUUAUUGCGCGUGUUGUGAUUUCGUGCUUUUAUGGUCCAUUUAA -----------------------.(((----(((((((...))))))).)))....(((((((.((((....))))..(((((...........)))))..)))))))........ ( -16.90) >DroPer_CAF1 37879 98 + 1 CUGUUG------------CUG--GAGC----CGACAACAUUGUUGUCAUAUUAGUAAACGUAACUAACUUUAGUUAUUGCUAGUGUUGCGACUUCGCACGUUUAUGGUCCAUUUAG ......------------.((--((.(----(((((((...))))))......((((((((....(((.(((((....))))).)))(((....)))))))))))))))))..... ( -28.70) >consensus ___________________UG__GAGC____CGACAACAUUGUUGUCAUAUUAGUAAACGUAACUAACUUUAGUUAUUGCUAGUGUUGCGACUUCGCACGUUUAUGGUCCAUUUAG ................................((((((...))))))(((((((((...(((((((....))))))))))))))))((((....)))).................. (-14.42 = -14.78 + 0.37)

| Location | 12,434,611 – 12,434,709 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 116 |
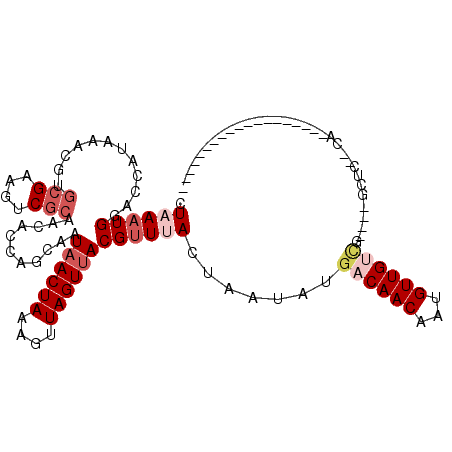
| Reading direction | reverse |
| Mean pairwise identity | 77.69 |
| Mean single sequence MFE | -20.12 |
| Consensus MFE | -8.88 |
| Energy contribution | -10.22 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12434611 98 - 23771897 CUAAAUGGACCAUAAACGUGCGAAGUCGCAACACCAGCAAUAACUAAAGUUAGUUACGUUUACUAAUAUGACAACAAUGUUGUUCUUGGGCCCUUCCC------------------ ......((.(((((((((((((....))))..........((((((....)))))))))))).......((((((...))))))..)))..)).....------------------ ( -20.00) >DroVir_CAF1 36949 89 - 1 UUAAAUGGACCAUAAAAGCGCGAAAUCGCAACACGCGCAAUAACUAAAGUUAGUAACGUUAACUAAUACAACAACAUUGUUGCCA----ACCC----------------------- .....(((.........(((((...........)))))..........((((((.......)))))).(((((....))))))))----....----------------------- ( -16.20) >DroPse_CAF1 37414 110 - 1 CUAAAUGGACCAUAAACGUGCGAAGUCGCAACACUAGCAAUAACUAAAGUUAGUUACGUUUACUAAUAUGACAACAAUGUUGUCG----GCUC--CAGCAUCAGCAUCAGCAACAG .....((((((.((((((((((....)))...((((((..........)))))).))))))).......((((((...)))))))----).))--))((....))........... ( -28.10) >DroYak_CAF1 39699 91 - 1 CUAAAUGGACCAUAAACGUGCGAAGUCGCAACACCAGCAAUAACUAAAGUUAGUUACGUUUACUAAUAUGACAACAAUGUUGUUC----------CACCAU--------------- ....((((....((((((((((....))))..........((((((....)))))))))))).......((((((...)))))).----------..))))--------------- ( -18.30) >DroMoj_CAF1 32921 89 - 1 UUAAAUGGACCAUAAAAGCACGAAAUCACAACACGCGCAAUAACUAAAGUUAGUUACGAUAACUAAUACGACAACAUUGUUGCCG----AUUC----------------------- .....(((.........((.((...........)).))..........((((((((...)))))))).(((((....))))))))----....----------------------- ( -11.20) >DroPer_CAF1 37879 98 - 1 CUAAAUGGACCAUAAACGUGCGAAGUCGCAACACUAGCAAUAACUAAAGUUAGUUACGUUUACUAAUAUGACAACAAUGUUGUCG----GCUC--CAG------------CAACAG .....((((((.((((((((((....)))...((((((..........)))))).))))))).......((((((...)))))))----).))--)).------------...... ( -26.90) >consensus CUAAAUGGACCAUAAACGUGCGAAGUCGCAACACCAGCAAUAACUAAAGUUAGUUACGUUUACUAAUAUGACAACAAUGUUGUCG____GCUC__CA___________________ .((((((............(((....)))...........((((((....)))))))))))).......((((((...))))))................................ ( -8.88 = -10.22 + 1.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:40 2006