
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,419,870 – 12,419,963 |
| Length | 93 |
| Max. P | 0.801854 |

| Location | 12,419,870 – 12,419,963 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 72.03 |
| Mean single sequence MFE | -21.75 |
| Consensus MFE | -10.95 |
| Energy contribution | -12.70 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
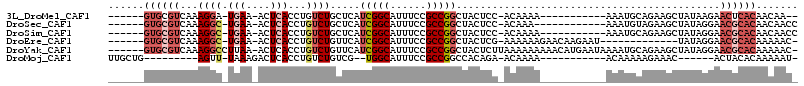
>3L_DroMel_CAF1 12419870 93 + 23771897 ------GUGCGUCAAAGGA-UGAA-ACUCACCUGUCUGCUCAUCGGCAUUUCCGCCGGCUACUCC-ACAAAA-----------AAAUGCAGAAGCUAUAAGAACUCACAACAA-- ------(((.(((..(((.-((..-...))))).(((((.....(((......)))((.....))-......-----------....)))))........).)).))).....-- ( -19.10) >DroSec_CAF1 22492 94 + 1 ------GUGCGUCAAAGGC-UGAA-ACUCACCUGUCUGCUCAUCGGCAUUUCCGCCGGCUACUCC-ACAAA------------AAAUGUAGAAGCUAUAGGAACGCACAACAACC ------((((((((.....-))).-.....(((((..(((..(((((......)))))((((...-.....------------....)))).))).)))))..)))))....... ( -25.50) >DroSim_CAF1 23108 95 + 1 ------GUGCGUCAAAGGC-UGAA-ACUCACCUGUCUGCUCAUCGGCAUUUCCGCCGGCUACUCC-ACAAAA-----------AAAUGCAGAAGCUAUAGGAACGCACAACAACC ------(((((((..(((.-((..-...))))).(((((.....(((......)))((.....))-......-----------....)))))........).))))))....... ( -24.70) >DroEre_CAF1 23451 92 + 1 ------GUGCGUCAAAGGC-UGAA-ACUCACCUGUCUGUUCAUCGGCAUUUCCGCCGGCUACUCG-AAAAAAGAACAAGAAU-------------UAUAGGAACGCACAAAAAC- ------((((((((.....-))).-.....(((((..((((.(((((......)))))....((.-......))....))))-------------.)))))..)))))......- ( -22.90) >DroYak_CAF1 24330 107 + 1 ------GUGCGUCAAAGGCCUUAA-ACUCACCUGUCUGUUCAUCGGCAUUUCCGCCGGCUACUCUUAAAAAAAAACAUGAAUAAAAUGCAGAAGCUAUAGGAACGCACAAAAAC- ------((((((...((.......-.))..(((((.(((((((((((......))))...................)))))))....((....)).))))).))))))......- ( -21.60) >DroMoj_CAF1 19735 84 + 1 UUGCUG---------AGUU-UAAAGACUCACCUGUCUGUCG--UGGCAUUUCCGCCGGCCACAGA-ACAAAA-----------ACAAAAAGAAAC------ACUACACAAAAAU- ....((---------((((-....))))))..(((((((.(--((((......))).)).)))).-)))...-----------............------.............- ( -16.70) >consensus ______GUGCGUCAAAGGC_UGAA_ACUCACCUGUCUGCUCAUCGGCAUUUCCGCCGGCUACUCC_ACAAAA___________AAAUGCAGAAGCUAUAGGAACGCACAAAAAC_ ......((((((...((((.(((....)))...)))).....(((((......)))))............................................))))))....... (-10.95 = -12.70 + 1.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:36 2006