| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,416,807 – 12,416,906 |
| Length | 99 |
| Max. P | 0.509177 |

| Location | 12,416,807 – 12,416,906 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 77.16 |
| Mean single sequence MFE | -27.08 |
| Consensus MFE | -11.38 |
| Energy contribution | -11.46 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.42 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
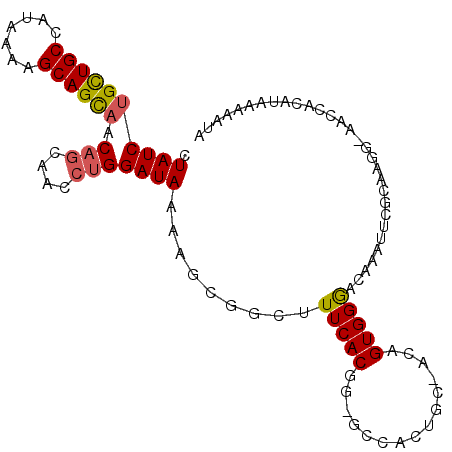
>3L_DroMel_CAF1 12416807 99 - 23771897 CUAUCUGCUGCCAUAAAAGCAGUAACAGCAACCUGGAUAAGAGCGGCUUUCACGA-GCCACUGC-ACAGUGGGACAGAUUCACAAUG-AAUCACAC-AAUUUA ..(((((.(.((((....(((((..(((....))).........(((((....))-))))))))-...)))).))))))........-........-...... ( -23.70) >DroPse_CAF1 19415 99 - 1 CUAUCUGUUGCCAUAAAAGCAGCCACAGCAACGCGGAUAAAAGCAGCUUUCACGG-AA--CUCCAACCGUGGAA-AGAUGCGACAUUAAAGUGGUUAAAAAUC ....(((((........)))))((((.((...))........(((.(((((((((-..--......)))).)))-)).))).........))))......... ( -23.20) >DroSim_CAF1 20032 100 - 1 CUAUCUGCUGCCAUAAAAGCAGCAACAGCAACCUGGAUAAGAGCGGCUUUCACGG-GCCACAGC-ACAGUGGGCCAAAUUCGCUAGG-AACAACAACAAAAUA .((((((((((.......)))))).(((....)))))))..((((..(((...((-.((((...-...)))).)))))..))))...-............... ( -27.10) >DroEre_CAF1 20449 100 - 1 CUAUCUGCUGCCAUAAAAGCAGCAACAGCAACCUGGAUAAAAGCGGCUUUCACGG-GCAGCUGC-ACAGUGGGCCAGAUUCGCCAGG-AGCCGCUUACAAAUA .((((((((((.......)))))).(((....))))))).(((((((((((((..-((....))-...)))(((.......))).))-))))))))....... ( -35.30) >DroYak_CAF1 21010 100 - 1 CUAUCUGCUGCCAUAAAAGCAGCAACAGCAACCUGGAUAAAAGCGGCUUUCACGG-GCAAGCGC-ACAGUGGGCCCAAUUCGCGAGG-AACCACAUAAAAAUA .((((((((((.......)))))).(((....)))))))...(.((.......((-((..((..-...))..))))..(((....))-).)).)......... ( -27.00) >DroAna_CAF1 15487 90 - 1 CUAUCUGCUGCCAUAAAAGCAGCAACAGCAACCUGGAUAAAAGCGGCUUUCACGCUGCCACAG--ACAGUGGGACAAGGUGGCA------CAAAGUAA----- .....((((((.......))))))...((.((((.(.....((((.......)))).((((..--...))))..).)))).)).------........----- ( -26.20) >consensus CUAUCUGCUGCCAUAAAAGCAGCAACAGCAACCUGGAUAAAAGCGGCUUUCACGG_GCCACUGC_ACAGUGGGACAAAUUCGCAAGG_AACCACAUAAAAAUA .((((((((((.......)))))).(((....))))))).........(((((...............))))).............................. (-11.38 = -11.46 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:34 2006