| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,399,416 – 12,399,507 |
| Length | 91 |
| Max. P | 0.932423 |

| Location | 12,399,416 – 12,399,507 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 79.37 |
| Mean single sequence MFE | -31.71 |
| Consensus MFE | -26.14 |
| Energy contribution | -26.37 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12399416 91 + 23771897 GGGUGGCCACCUUCGGCUUGGAGCCGCCAAUUAGGCCAGGUGGUCGCAGGGUGCCUGCAAGAUGGAUA-----AAUUGGGGUUUCAACAA--UGUAC----U .(((((((......)))((((((((.((((((.((((....))))(((((...)))))..........-----))))))))))))))...--..)))----) ( -33.70) >DroSec_CAF1 2029 93 + 1 GGGUGGCCACCUUCGGCUUGGAGCCGCCAAUUAGGCCAGGUGGUCGCAGGGUGCCUGUAAGAUGGAUA-----AAUUGGGGGUUAAACUAUAUAUAC----U ..((((((((((..(((.....)))(((.....))).))))))))))((...((((.(((........-----..))).))))....))........----. ( -32.20) >DroSim_CAF1 2008 91 + 1 GGGUGGCCACCUUCGGCUUGGAGCCGCCAAUUAGGCCAGGUGGUCGCAGGGUGCCUGCAAGAUGGAUA-----AAUUGGGGGUUAAGCAU--UGUAC----U ..((((((((((..(((.....)))(((.....))).))))))))))...((((((.(((........-----..))).))))...))..--.....----. ( -32.90) >DroEre_CAF1 1958 70 + 1 GGGUGGCCACCUUCGGCUUGGAGCCGCCAAUUAGGCCAGGUGGUCGCAGGGUGCCUGCAAGAUGGAUA-----AA--------------------------- ....((((((((..(((.....)))(((.....))).))))))))(((((...)))))..........-----..--------------------------- ( -29.70) >DroYak_CAF1 2055 100 + 1 GGGUGGCCACCUUCGGCUUGGAGCCGCCAAUUAGGCCAGGUGGUCGCAGGGUGCCUGCAAGAUGGAUAUGGUAAAUUGCGGGUUAGACAU--GGUACAUGGU ..((((((((((..(((.....)))(((.....))).)))))))))).....((((((((...............))))))))....(((--(...)))).. ( -37.46) >DroAna_CAF1 1683 91 + 1 GGGUGGCCACUUUGGGCUUGGAGCCUCCUAUUAGGCCCGGAGGUCGCAGCGUGCCUGCAAAGAAAAGA-----GAUUGUGGAUUGAAACU--UGUAU----U ..((((((..(((((((((((((....)).))))))))))))))))).......(..(((........-----..)))..).........--.....----. ( -24.30) >consensus GGGUGGCCACCUUCGGCUUGGAGCCGCCAAUUAGGCCAGGUGGUCGCAGGGUGCCUGCAAGAUGGAUA_____AAUUGGGGGUUAAACAU__UGUAC____U ....((((((((..(((.....)))(((.....))).))))))))((((.....))))............................................ (-26.14 = -26.37 + 0.22)

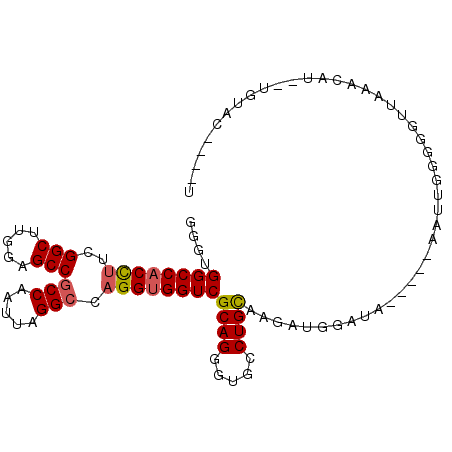
| Location | 12,399,416 – 12,399,507 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 79.37 |
| Mean single sequence MFE | -25.62 |
| Consensus MFE | -22.30 |
| Energy contribution | -23.13 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12399416 91 - 23771897 A----GUACA--UUGUUGAAACCCCAAUU-----UAUCCAUCUUGCAGGCACCCUGCGACCACCUGGCCUAAUUGGCGGCUCCAAGCCGAAGGUGGCCACCC .----.....--..((((......)))).-----........(((((((...)))))))((((((.(((.....)))(((.....)))..))))))...... ( -27.70) >DroSec_CAF1 2029 93 - 1 A----GUAUAUAUAGUUUAACCCCCAAUU-----UAUCCAUCUUACAGGCACCCUGCGACCACCUGGCCUAAUUGGCGGCUCCAAGCCGAAGGUGGCCACCC .----........................-----...........((((...)))).(.((((((.(((.....)))(((.....)))..)))))).).... ( -22.80) >DroSim_CAF1 2008 91 - 1 A----GUACA--AUGCUUAACCCCCAAUU-----UAUCCAUCUUGCAGGCACCCUGCGACCACCUGGCCUAAUUGGCGGCUCCAAGCCGAAGGUGGCCACCC .----.....--.................-----........(((((((...)))))))((((((.(((.....)))(((.....)))..))))))...... ( -27.40) >DroEre_CAF1 1958 70 - 1 ---------------------------UU-----UAUCCAUCUUGCAGGCACCCUGCGACCACCUGGCCUAAUUGGCGGCUCCAAGCCGAAGGUGGCCACCC ---------------------------..-----........(((((((...)))))))((((((.(((.....)))(((.....)))..))))))...... ( -27.40) >DroYak_CAF1 2055 100 - 1 ACCAUGUACC--AUGUCUAACCCGCAAUUUACCAUAUCCAUCUUGCAGGCACCCUGCGACCACCUGGCCUAAUUGGCGGCUCCAAGCCGAAGGUGGCCACCC ..((((...)--)))...........................(((((((...)))))))((((((.(((.....)))(((.....)))..))))))...... ( -27.90) >DroAna_CAF1 1683 91 - 1 A----AUACA--AGUUUCAAUCCACAAUC-----UCUUUUCUUUGCAGGCACGCUGCGACCUCCGGGCCUAAUAGGAGGCUCCAAGCCCAAAGUGGCCACCC .----.....--.........((((....-----........((((((.....)))))).....((((((......)))).)).........))))...... ( -20.50) >consensus A____GUACA__AUGUUUAACCCCCAAUU_____UAUCCAUCUUGCAGGCACCCUGCGACCACCUGGCCUAAUUGGCGGCUCCAAGCCGAAGGUGGCCACCC ..........................................((((((.....))))))((((((.(((.....)))(((.....)))..))))))...... (-22.30 = -23.13 + 0.83)

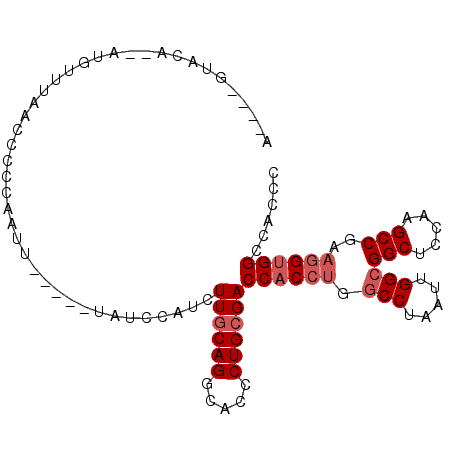
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:27 2006