| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,376,930 – 12,377,021 |
| Length | 91 |
| Max. P | 0.866645 |

| Location | 12,376,930 – 12,377,021 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 81.24 |
| Mean single sequence MFE | -18.86 |
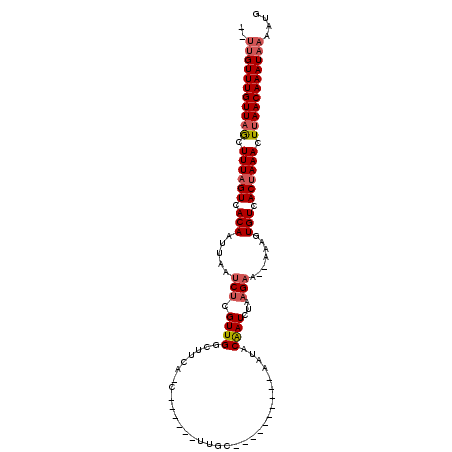
| Consensus MFE | -13.39 |
| Energy contribution | -14.00 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866645 |
| Prediction | RNA |
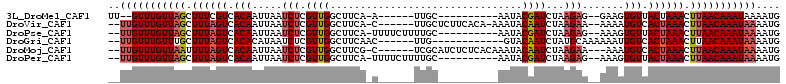
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12376930 91 + 23771897 UU--GUUUGUUAGCUUUCGUCACAAUUAAUCUCGUUGGCUUCA-A------UUGC----------AAUACGAUCUAAGAG--GAAGUGUUACUAAACUUAACAAAUAAAAUG ((--(((((((((.(((.((.(((.((..((((((..((....-.------..))----------...)))).....)).--.)).))).)).))).))))))))))).... ( -18.20) >DroVir_CAF1 3433 100 + 1 --UUGUUUGUUAGCUUUAGUCACAAUUAAUCUCGUUGGCUUCA-C------UUGCUCUUCACA-AAAUACAAUCUAAGAA--AAAAUGUCACUAAACUUAACAAAUAAAAUG --(((((((((((.((((((.(((.((..(((.((((......-.------............-.....))))...))).--.)).))).)))))).))))))))))).... ( -16.96) >DroPse_CAF1 3502 97 + 1 --UUGUUUGUUAGCUUUAGUCACAAUUAAUCUCGUUGGCUUCA-UUUUCUUUUGC----------AAUACGAUCUAAGAG--AAAGUGUUACUAAACUUAACAAAUAAAAUG --(((((((((((.((((((..((((.......))))....((-((((((((((.----------.........))))))--))))))..)))))).))))))))))).... ( -22.40) >DroGri_CAF1 3437 92 + 1 --UUGUUUGUUUGCUUUAGUCACACAUAAUCUCGUUGGCUUCAAC------UUG------------GUACAAUCUAUGCAAAAAAUUGUCACUAAACUUAACAAAUAAAAUG --(((((((((...((((((.(((.........((((....))))------.((------------(......)))..........))).))))))...))))))))).... ( -15.30) >DroMoj_CAF1 3462 100 + 1 --UUGUUUGUUAAUUUUAGUCACAAUUAAUCUCGUUGGCUUCG-C------UCGCAUCUCUCACAAAUACAAUCUAAGAA---AAAUGUCACUAAACUUAACAAAUAAAAUG --(((((((((((.((((((.(((.........((.(((...)-)------).)).(((.................))).---...))).)))))).))))))))))).... ( -17.93) >DroPer_CAF1 3503 97 + 1 --UUGUUUGUUAGCUUUAGUCACAAUUAAUCUCGUUGGCUUCA-UUUUCUUUUGC----------AAUACGAUCUAAGAG--AAAGUGUUACUAAACUUAACAAAUAAAAUG --(((((((((((.((((((..((((.......))))....((-((((((((((.----------.........))))))--))))))..)))))).))))))))))).... ( -22.40) >consensus __UUGUUUGUUAGCUUUAGUCACAAUUAAUCUCGUUGGCUUCA_C______UUGC__________AAUACAAUCUAAGAA__AAAGUGUCACUAAACUUAACAAAUAAAAUG ..(((((((((((.((((((.(((.....(((.((((................................))))...))).......))).)))))).))))))))))).... (-13.39 = -14.00 + 0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:18 2006