| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,370,708 – 12,370,810 |
| Length | 102 |
| Max. P | 0.642181 |

| Location | 12,370,708 – 12,370,810 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 78.62 |
| Mean single sequence MFE | -22.47 |
| Consensus MFE | -17.12 |
| Energy contribution | -16.90 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
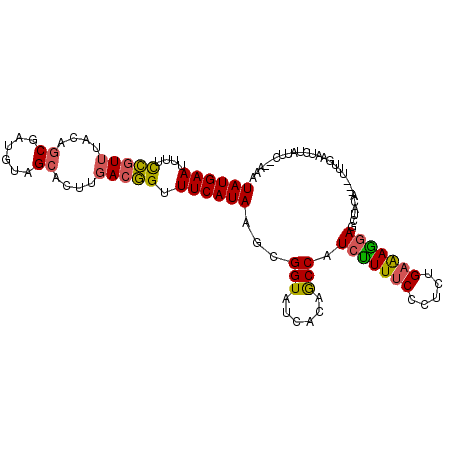
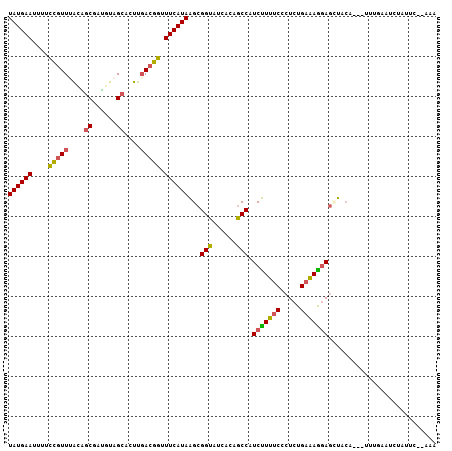
>3L_DroMel_CAF1 12370708 102 - 23771897 UAUGAAUUUUCCGUUUACAGCAAUGUAGCACUUGACGGUUUCAUAAGCGGUAUCACAGCCAUCUUUUCCCUCUGAAAAGAGCUACA---UUUGAGUUUGUUC--UAA ((((((....(((((((((....))))......))))).))))))((((((......))).(((((((.....))))))))))...---.............--... ( -23.50) >DroSec_CAF1 55853 102 - 1 UAUGAAUUUUCCGUUUACAGCGAUGUAGCACUUGACGGUUUCAUAAGCGGUUUCACAGCCAUCUUUUCCCUCUGAAAGGAGAUACA---UUUUAAUCUAUUC--AAA ((((((....(((((....((......))....))))).))))))...(((......))).........((((....)))).....---.............--... ( -20.80) >DroSim_CAF1 59300 102 - 1 UAUGAAUUUUCCGUUUACAGCAAUGUAGCACUUGACGGUUUCAUAAGCGGUUUCACAACCAUCUUUUCCCUCUGAAAGGAGAUACA---UUUUAAUCAGUUC--GAA ((((((....(((((((((....))))......))))).))))))(((((((....)))).........((((....)))).....---.........))).--... ( -20.30) >DroEre_CAF1 55918 107 - 1 UAUGAAUUUGCCGUUUGUAGCGAUGUAGCAUUUGACGGUUUCAUAAGCGGUAUCACAGCCAUCUUUUCCCUCUGAGAAGAGCUCCAAAUUGUGAUUUUAUUAUUAUG ((((((...((((((....((......))....))))))))))))((((((......))).(((((((.....)))))))))).......(((((......))))). ( -25.20) >DroYak_CAF1 56614 104 - 1 UAUGAAUUUUCCGUUGACGGCGAUGUAGCACUUGACGGUUUCAUAAGCGGUAUCACAGCCAUCUUUGCCCUCUGAAAGGAGCUUCAAAAUGUGAUUUCAUU---AUA ((((((....((((..(..((......))..)..)))).))))))...((.((((((......((((..((((....))))...)))).)))))).))...---... ( -26.90) >DroAna_CAF1 51765 103 - 1 UAUGAAUGGUUUAUUAACCACCAGAUAGCACUUUACAGUUUCAUAGGCGGUUUCACAGCCAUCCUUUCCCUCUGAAAGUAAUUCGAGAUUCACGGACAGUUC--U-- ..(((((((((....))))....((....(((((.(((......(((.(((......)))..)))......))))))))...))...)))))..........--.-- ( -18.10) >consensus UAUGAAUUUUCCGUUUACAGCGAUGUAGCACUUGACGGUUUCAUAAGCGGUAUCACAGCCAUCUUUUCCCUCUGAAAGGAGCUACA___UUUGAAUCUAUUC__AAA ((((((....(((((....((......))....))))).))))))...(((......))).(((((((.....)))))))........................... (-17.12 = -16.90 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:15 2006