| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,364,484 – 12,364,596 |
| Length | 112 |
| Max. P | 0.719888 |

| Location | 12,364,484 – 12,364,596 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.96 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -10.88 |
| Energy contribution | -11.24 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
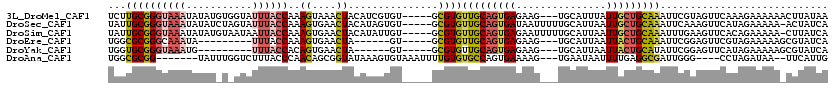
>3L_DroMel_CAF1 12364484 112 - 23771897 UCUUGCGGGUAAAUAUAUGUGGUAUUUACCAAAGUAAACUACAUCGUGU-----GCGUGUUGCAGUGAGAAG---UGCAUUUAUUGCUGCAAAUUCGUAGUUCAAAGAAAAAACUUAUAA ((((((.(((((((((.....)))))))))...)).((((((..((...-----.))..((((((..((((.---....))).)..))))))....))))))..))))............ ( -26.60) >DroSec_CAF1 49562 114 - 1 UAUUGCGGGUAAAUAUAUCUAGUAUUUACCAAAGUGAACUACAUAGUGU-----GCGUGUUGCAGUGAUAAUUUUUGCAUUAAUUGCUGCAAAUUCAAAGUUCAUAGAAAAA-ACUAUCA .......(((((((((.....)))))))))...(((((((((.....))-----..(..((((((..(((((......))).))..))))))...)..))))))).......-....... ( -25.90) >DroSim_CAF1 53022 114 - 1 UAUUGCGGGUAAAUAUAUGUAAUAAUUACCAAAGUGAACUACAUAUUGU-----GCGUGUUGCAGUGAGAAUUUUUGCAUUAAUUGCUGCAAAUUUGAAGUUCACAGAAAAA-CUUAUCA .......(((((.(((.....))).)))))...(((((((.((..((((-----(((...(((((.........))))).....))).))))...)).))))))).......-....... ( -24.30) >DroEre_CAF1 49767 97 - 1 UGGCGCGGGCAAAUA---------UUUACCAAAGUGAACUA------GU-----GCGUGUUGCAGUGAGAAG---UGCAUUAAUUACUGCAAAUUCGGAGUUCGUAGAAAAAGCGUAUCA ..((((((.......---------....)).....(((((.------..-----.((..(((((((((....---........)))))))))...)).))))).........)))).... ( -24.40) >DroYak_CAF1 50148 97 - 1 UGGUGCGGGUAAAUG---------UUUACCACAGUGAACUA------GU-----GCGUGUUGCAGUGAGAAG---UGCAUUAAUUACUGCAUAUUCGGAGUUCAUAGAAAAAGCGUAUCA .(((((((((((...---------.)))))...(((((((.------.(-----(.((((.(((((((....---........))))))))))).)).)))))))........)))))). ( -29.70) >DroAna_CAF1 46421 104 - 1 UGGCGCGG-------UAUUUGGUCUUUACCCAACAGCGGUAUAAAGUGUAAAUUUUGUGUGCCAGUGAAAAG---UGAAUAAUUUUGAGGCGAUUGGG----CCUAGAUAA--UUCAUUG ((((((((-------((.........))))..........(((((((....)))))))))))))......((---(((((.(((...((((......)----))).))).)--)))))). ( -26.80) >consensus UGGUGCGGGUAAAUAUAU_U_GUAUUUACCAAAGUGAACUACAUAGUGU_____GCGUGUUGCAGUGAGAAG___UGCAUUAAUUGCUGCAAAUUCGGAGUUCAUAGAAAAA_CUUAUCA ...((((((((((...........))))))..((....))...............))))(((((((((...............)))))))))............................ (-10.88 = -11.24 + 0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:14 2006