| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,361,195 – 12,361,299 |
| Length | 104 |
| Max. P | 0.993385 |

| Location | 12,361,195 – 12,361,299 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 80.81 |
| Mean single sequence MFE | -27.35 |
| Consensus MFE | -17.81 |
| Energy contribution | -18.12 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12361195 104 + 23771897 AACCCAAAACCAUUUACCUGGCCAAUUGGCCAUGGCUUUUUGUGUUCUUCACUGUU--CAGUUUGGGCCUGUUCUCUGCCGUUCUGUGCGGUUCUGGCCAAACAAA ..((((((........(((((((....))))).))......(((.....)))....--...))))))..((..(...(((((.....)))))...)..))...... ( -28.10) >DroSec_CAF1 46172 92 + 1 AACCCAAAACCAUUUACCUGGCCAAUUGGCCAUGGCUUUCU--GUUCUUCACUGUU--CAGUUUGGGCCUGUUCU----------GUGCGGUUCUGGCCAAACAAA ...(((.((((.......(((((....))))).(((((.((--(..(......)..--)))...)))))......----------....)))).)))......... ( -26.50) >DroSim_CAF1 49405 104 + 1 AACCCAAAACCAUUUACCUGGCCAAUUGGCCAUGGCUUGUG--UUUCUUCACUGUUCUCAGUUUGGGCCUGUUCUGUGCCGUUCUGUGCGGUUCUGGCCAAACAAA ....(((...(((...(((((((....))))).))...)))--.......((((....)))))))((((.(..(((..(......)..)))..).))))....... ( -30.60) >DroYak_CAF1 46818 90 + 1 AACCCAAAACCAUUUACCUGGCCAAUUGGCCAUGGCUUUUG--GUUCUUCACUCUU--AAUUUCUGGUCAGAUU-----UGUUCUGUUCUGUUCUGGCC------- .......(((((....(((((((....))))).))....))--)))..........--.......(((((((..-----.(.......)...)))))))------- ( -24.20) >consensus AACCCAAAACCAUUUACCUGGCCAAUUGGCCAUGGCUUUUG__GUUCUUCACUGUU__CAGUUUGGGCCUGUUCU____CGUUCUGUGCGGUUCUGGCCAAACAAA ...(((.((((.......(((((....))))).(((((..........................)))))....................)))).)))......... (-17.81 = -18.12 + 0.31)

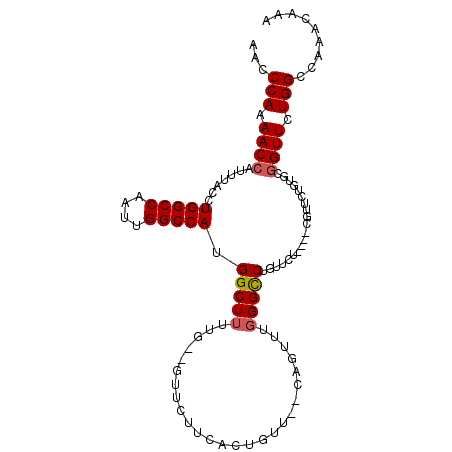

| Location | 12,361,195 – 12,361,299 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 80.81 |
| Mean single sequence MFE | -29.64 |
| Consensus MFE | -24.27 |
| Energy contribution | -24.40 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12361195 104 - 23771897 UUUGUUUGGCCAGAACCGCACAGAACGGCAGAGAACAGGCCCAAACUG--AACAGUGAAGAACACAAAAAGCCAUGGCCAAUUGGCCAGGUAAAUGGUUUUGGGUU .((((((.(((...............)))...))))))(((((((((.--....(((.....))).....(((.(((((....))))))))....)).))))))). ( -31.56) >DroSec_CAF1 46172 92 - 1 UUUGUUUGGCCAGAACCGCAC----------AGAACAGGCCCAAACUG--AACAGUGAAGAAC--AGAAAGCCAUGGCCAAUUGGCCAGGUAAAUGGUUUUGGGUU .........((((((((((((----------....(((.......)))--....)))......--.....(((.(((((....))))))))...)))))))))... ( -29.20) >DroSim_CAF1 49405 104 - 1 UUUGUUUGGCCAGAACCGCACAGAACGGCACAGAACAGGCCCAAACUGAGAACAGUGAAGAAA--CACAAGCCAUGGCCAAUUGGCCAGGUAAAUGGUUUUGGGUU .........(((((((((..(((...(((.(......))))....)))......(((......--)))..(((.(((((....))))))))...)))))))))... ( -32.10) >DroYak_CAF1 46818 90 - 1 -------GGCCAGAACAGAACAGAACA-----AAUCUGACCAGAAAUU--AAGAGUGAAGAAC--CAAAAGCCAUGGCCAAUUGGCCAGGUAAAUGGUUUUGGGUU -------.(((.........((((...-----..))))..........--........(((((--((...(((.(((((....))))))))...))))))).))). ( -25.70) >consensus UUUGUUUGGCCAGAACCGCACAGAACG____AGAACAGGCCCAAACUG__AACAGUGAAGAAC__AAAAAGCCAUGGCCAAUUGGCCAGGUAAAUGGUUUUGGGUU .........(((((((((..........................((((....))))..............(((.(((((....))))))))...)))))))))... (-24.27 = -24.40 + 0.13)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:11 2006