| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,339,002 – 12,339,110 |
| Length | 108 |
| Max. P | 0.969379 |

| Location | 12,339,002 – 12,339,110 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 75.71 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -11.18 |
| Energy contribution | -12.54 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
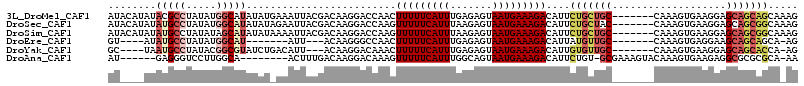
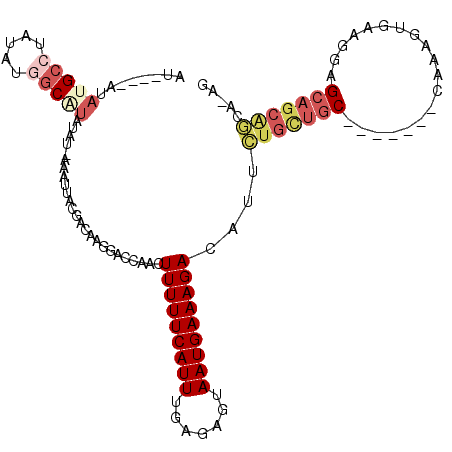
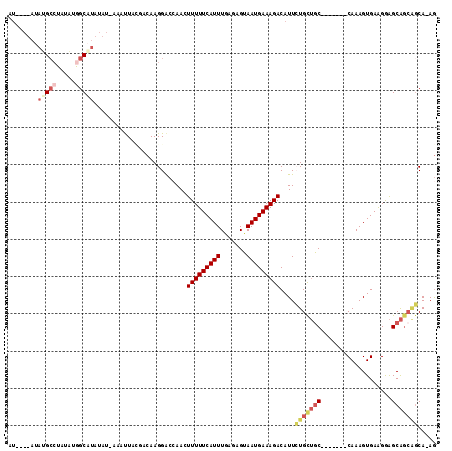
>3L_DroMel_CAF1 12339002 108 + 23771897 AUACAUAUACGCCUAUAUGGCAUAUAUGAAAUUACGACAAGGACCAACUUUUUCAUUUGAGAGUAAUGAAAGACAUUCUGCUGC-------CAAAGUGAAGGAGCAGCAGCAAAG ...((((((.(((.....))).))))))....................((((((((((....).)))))))))....(((((((-------(.........).)))))))..... ( -26.20) >DroSec_CAF1 24150 108 + 1 AUACAUAUAUGCCUAUAUGGCAUAUAUAGAAUUACGACAAGGACCAAGUUUUUCAUUUAAGAGUAAUGAAAGACAUUCUGCUAC-------CAAAGUGAAGGAGCAGCGGCAAAG ....(((((((((.....)))))))))..............(.((..((((((((((.......))))))))))...(((((.(-------(........))))))).))).... ( -30.80) >DroSim_CAF1 24780 108 + 1 AUACAUAUAUGCCUAUAUAGCAUAUAUAAAAUUACGACAAGGACCAAGUUUUUCAUUUAAGAGUAAUGAAAGACAUUCUGCUGC-------CAAAGUGAAGGAGCAGCGGCAAAG ....((((((((.......))))))))..............(.((..((((((((((.......))))))))))...(((((.(-------(........))))))).))).... ( -26.20) >DroEre_CAF1 24349 93 + 1 GU----AUAUGCCUAUAUGGCAU-------AUU---ACAAGGGCCAACUUUUUCAUUUGAGAGUAAUGAAAGACAUUAUGUUGC-------CAAAGUGAGGAAGCAGCAGCA-AG ((----(((((((.....)))))-------).)---))....((...(((((((((((..(.((((((.....))))))....)-------..)))))))))))..))....-.. ( -25.30) >DroYak_CAF1 24400 100 + 1 GC----UAAUGCCUAUACGGCGUAUCUGACAUU---ACAAGGACAAACUUUUUCAUUUGAGAGUAAUGAAAGACAUUGUGUUGC-------CAAAGUGAAGGAGCAGCACCA-AG ..----..(((((.....))))).(((..((((---(((((......)))((((....))))))))))..)))....(((((((-------(.........).)))))))..-.. ( -23.50) >DroAna_CAF1 23034 99 + 1 AU------GAGGGUCCUUGGCA--------ACUUUGACAAGGACAAAGUUUUUCAUUUGGCAGUAAUGAAAGACAUUCUGU-GCGAAAGUACAAAGUGAAGAGGCGCGCGCA-AA ..------(((.(((((((.((--------....)).)))))))...((((((((((.......)))))))))).)))(((-((....)))))..(((......))).....-.. ( -30.00) >consensus AU____AUAUGCCUAUAUGGCAUAUAU_AAAUUACGACAAGGACCAACUUUUUCAUUUGAGAGUAAUGAAAGACAUUCUGCUGC_______CAAAGUGAAGGAGCAGCAGCA_AG ........(((((.....))))).........................(((((((((.......)))))))))....(((((((...................)))))))..... (-11.18 = -12.54 + 1.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:59 2006