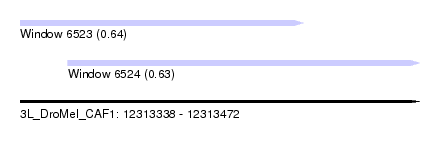
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,313,338 – 12,313,472 |
| Length | 134 |
| Max. P | 0.638449 |

| Location | 12,313,338 – 12,313,433 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 90.70 |
| Mean single sequence MFE | -20.78 |
| Consensus MFE | -18.67 |
| Energy contribution | -18.18 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12313338 95 + 23771897 GUCAUAUAAUUGUAAAUUGUAACUGACUCUGAUUGCACAUUGAUUAUAAUUGUAAUUAUUGUUGCUUAUAUCGGAUUACACGCUGGCGAAUUGGC ((((((((((.....))))))..))))((((((.(((((.(((((((....))))))).)).)))....))))))......((..(....)..)) ( -19.90) >DroSec_CAF1 48463 95 + 1 GUAAUUUAAGCUGAAAUUGUAAGUGACUCUGAUUGCACAUUGAUUAUAAUUGUAAUUAUUGUUGCUUAUAUCGGAUUACACGCUGGCGAAUUCGC (((((.....((((...((((((..((..((((((((.((((....))))))))))))..))..)))))))))))))))......(((....))) ( -22.80) >DroSim_CAF1 48342 95 + 1 GUCAUUUAAGCUGAAAUUGUAAGUGACUCUGAUUGCACAUUGAUUAUAAUUGUAAUUAUUGUUGCUUAUAUCGGGUUACACGCUGGCGAAUUCGC ((((..(((.((((...((((((..((..((((((((.((((....))))))))))))..))..)))))))))).))).....))))........ ( -23.70) >DroYak_CAF1 52075 94 + 1 GUCAUUUAAGCUGAAAUUGUAAGUGACUCUGAUUGCACACUGAUUAUAAUUGUAUUUAUUGUAGCUUAUAUCAGAUUACAUA-UAGCGAAUUUGC .........((((....(((((.....((((((.(((((.(((.(((....))).))).))).))....)))))))))))..-))))........ ( -16.70) >consensus GUCAUUUAAGCUGAAAUUGUAAGUGACUCUGAUUGCACAUUGAUUAUAAUUGUAAUUAUUGUUGCUUAUAUCGGAUUACACGCUGGCGAAUUCGC ............(((.((((..(((..((((((.(((((.(((((((....))))))).))).))....))))))...)))....)))).))).. (-18.67 = -18.18 + -0.50)


| Location | 12,313,354 – 12,313,472 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 88.28 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -19.10 |
| Energy contribution | -19.48 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12313354 118 + 23771897 UUGUAACUGACUCUGAUUGCACAUUGAUUAUAAUUGUAAUUAUUGUUGCUUAUAUCGGAUUACACGCUGGCGAAUUGGCAACAUUUGAAACAUUCAGAAGUUAAAUCUAUUUUUUUCA .(((((.....((((((.(((((.(((((((....))))))).)).)))....))))))))))).((..(....)..))(((.(((((.....))))).)))................ ( -22.20) >DroSec_CAF1 48479 118 + 1 UUGUAAGUGACUCUGAUUGCACAUUGAUUAUAAUUGUAAUUAUUGUUGCUUAUAUCGGAUUACACGCUGGCGAAUUCGCAACAUUUGAAACAUUAUAAAGUUAAAUCUAUUUUCUUCA ((((((((((.((((((.(((((.(((((((....))))))).)).)))....))))))))))......(((....))).............)))))).................... ( -23.00) >DroSim_CAF1 48358 118 + 1 UUGUAAGUGACUCUGAUUGCACAUUGAUUAUAAUUGUAAUUAUUGUUGCUUAUAUCGGGUUACACGCUGGCGAAUUCGCAACAUUUGAAACAUUCAAAAGUGAAAUCUAUUUUCUUCA .((((((..((..((((((((.((((....))))))))))))..))..))))))..(((((.(((....(((....)))....(((((.....))))).))).))))).......... ( -28.90) >DroYak_CAF1 52091 114 + 1 UUGUAAGUGACUCUGAUUGCACACUGAUUAUAAUUGUAUUUAUUGUAGCUUAUAUCAGAUUACAUA-UAGCGAAUUUGCAACAUUUGUAAUUUGCAGACGUUAAACCUAUAC---UCA ..(((.((((.((((((.(((((.(((.(((....))).))).))).))....))))))))))...-(((((..(((((((..........))))))))))))......)))---... ( -23.10) >consensus UUGUAAGUGACUCUGAUUGCACAUUGAUUAUAAUUGUAAUUAUUGUUGCUUAUAUCGGAUUACACGCUGGCGAAUUCGCAACAUUUGAAACAUUCAAAAGUUAAAUCUAUUUUCUUCA ......((((.((((((.(((((.(((((((....))))))).))).))....))))))))))......(((....)))(((.(((((.....))))).)))................ (-19.10 = -19.48 + 0.37)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:51 2006