
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,305,512 – 12,305,655 |
| Length | 143 |
| Max. P | 0.979489 |

| Location | 12,305,512 – 12,305,623 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.95 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -16.06 |
| Energy contribution | -17.28 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12305512 111 - 23771897 CGAGCGGAAACGGAAACUAUUUGCUGCCAUU-UGG-GCAAUGGCAAAUGGCAAAUAGCAAAUGGCAGAAUGCGAGCAUUAAUAGAAAUUUCAUAGGCAAAUGCAAAGUUGGCC ....((....))(((((((((((((((((((-((.-.......))))))))....)))))))))...((((....))))........))))...(((.(((.....))).))) ( -30.10) >DroSim_CAF1 40525 107 - 1 ----CGGAAACGGAAACUAUUUGCUGCCAUU-UGG-GCAAUGGCAAAUGGCAAAUAGCAAAUGGCAGAAUGCGAGCAUUAAUAGAAAUUUCAUAGGCAAACGCAAAGUUGGCC ----((....))(((((((((((((((((((-((.-.......))))))))....)))))))))...((((....))))........))))...(((.(((.....))).))) ( -32.10) >DroEre_CAF1 41232 106 - 1 ----CGGAAACGGAAACUAUUUGCUGCCAUU-UGG-GCAAUGGCAAACGGCAA-UGGCAAGUGGCAGAAUGCGAGCAUUAAUAGAAAUUUCAUAGGCAAAUGCUGAGUUGGCC ----((....))(((((((((..((((((((-(.(-.((.((.(....).)).-)).))))))))))((((....)))))))))...))))...(((.(((.....))).))) ( -29.20) >DroWil_CAF1 60105 94 - 1 ----CGGAAACGGAAACUAUUUGCUGGCAUU-UCA-G-------------CAAAUGGCAAAUGGCAAAAGCCAAACAUUAAUAGAAAUUUCAUAUGCAAAUAUAAAGUUUAGC ----((....))....(((((((((((....-)))-)-------------)))))))....((((....))))...........((((((.((((....)))).))))))... ( -24.40) >DroYak_CAF1 43384 104 - 1 ----CGGAAACGGAAACUAUUUGCUGCCAUUUUGG-GCAAUGGCAAGUGGCAAGUGGCAAAUGGCAGAAUGCGAGCAUUAAUAGAAAUUUCAUAGGCAAAUGCAAAGUU---- ----((....))(((((((((..((((((((...(-.((...((.....))...)).).))))))))((((....)))))))))...))))....((....))......---- ( -26.00) >DroAna_CAF1 37510 104 - 1 ----CGGAAGCGGAAACUAUUUGCUGCCAUU-UGGAUCAAUGGCGGA--GCAAAUGGUAAAUGGCAAGCGGCAAGUAUUAAUAGAAAUUUCAUUGGAAGCCCCAGAUCUAG-- ----.((..((.(..((((((((((((((((-......))))))..)--)))))))))...).))....(((.....(((((.((....)))))))..)))))........-- ( -25.00) >consensus ____CGGAAACGGAAACUAUUUGCUGCCAUU_UGG_GCAAUGGCAAAUGGCAAAUGGCAAAUGGCAGAAUGCGAGCAUUAAUAGAAAUUUCAUAGGCAAAUGCAAAGUUGGCC ....((....))(((((((((((((((((((.......)))))))....))))))))......((.....))...............))))...................... (-16.06 = -17.28 + 1.23)

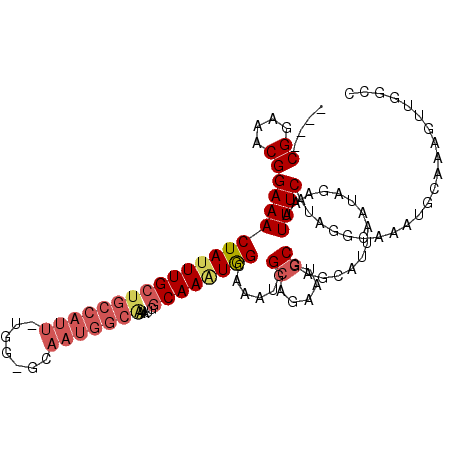

| Location | 12,305,546 – 12,305,655 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.94 |
| Mean single sequence MFE | -19.53 |
| Consensus MFE | -15.52 |
| Energy contribution | -16.32 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12305546 109 + 23771897 AAUGCUCGCAUUCUGCCAUUUGCUAUUUGCCAUUUGCCAUUGCCCA-AAUGGCAGCAAAUAGUUUCCGUUUCCGCUCGUUCCGUGCCACUACCCCAAUCCACACACCACU ...((.((......((.....(((((((((....(((((((.....-))))))))))))))))..........))......)).))........................ ( -21.76) >DroSec_CAF1 40739 90 + 1 AAUGCUCGCAUUCUGCCAUUUGCUAUUUGCCAUUUGCCAUUGCCCA-AAUGGCAGCAAAUAGUUUCCGUUUCCG-------------------CCAAUCCACACACCCCU ...((..((.....)).....(((((((((....(((((((.....-))))))))))))))))..........)-------------------)................ ( -20.50) >DroSim_CAF1 40559 90 + 1 AAUGCUCGCAUUCUGCCAUUUGCUAUUUGCCAUUUGCCAUUGCCCA-AAUGGCAGCAAAUAGUUUCCGUUUCCG-------------------CCAAUCCACACACCCCU ...((..((.....)).....(((((((((....(((((((.....-))))))))))))))))..........)-------------------)................ ( -20.50) >DroEre_CAF1 41266 88 + 1 AAUGCUCGCAUUCUGCCACUUGCCA-UUGCCGUUUGCCAUUGCCCA-AAUGGCAGCAAAUAGUUUCCGUUUCCG-------------------CUGCGCCGUGCCACAA- .......((((..(((...((((..-.(((((((((........))-)))))))))))..(((..........)-------------------)))))..)))).....- ( -18.80) >DroYak_CAF1 43414 91 + 1 AAUGCUCGCAUUCUGCCAUUUGCCACUUGCCACUUGCCAUUGCCCAAAAUGGCAGCAAAUAGUUUCCGUUUCCG-------------------CUUCUCCCUGCCGCCCC ..(((..(((....((.....))....)))....(((((((......))))))))))................(-------------------(........))...... ( -16.10) >consensus AAUGCUCGCAUUCUGCCAUUUGCUAUUUGCCAUUUGCCAUUGCCCA_AAUGGCAGCAAAUAGUUUCCGUUUCCG___________________CCAAUCCACACACCCCU .......((.....)).....(((((((((....(((((((......))))))))))))))))............................................... (-15.52 = -16.32 + 0.80)



| Location | 12,305,546 – 12,305,655 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 81.94 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -22.96 |
| Energy contribution | -22.52 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12305546 109 - 23771897 AGUGGUGUGUGGAUUGGGGUAGUGGCACGGAACGAGCGGAAACGGAAACUAUUUGCUGCCAUU-UGGGCAAUGGCAAAUGGCAAAUAGCAAAUGGCAGAAUGCGAGCAUU .((..(((((.(..........).)))))..))...((....))....(((((((((((((((-((........))))))))....)))))))))...((((....)))) ( -30.00) >DroSec_CAF1 40739 90 - 1 AGGGGUGUGUGGAUUGG-------------------CGGAAACGGAAACUAUUUGCUGCCAUU-UGGGCAAUGGCAAAUGGCAAAUAGCAAAUGGCAGAAUGCGAGCAUU ...(((((.((.(((..-------------------((....))....(((((((((((((((-((........))))))))....)))))))))...))).)).))))) ( -28.80) >DroSim_CAF1 40559 90 - 1 AGGGGUGUGUGGAUUGG-------------------CGGAAACGGAAACUAUUUGCUGCCAUU-UGGGCAAUGGCAAAUGGCAAAUAGCAAAUGGCAGAAUGCGAGCAUU ...(((((.((.(((..-------------------((....))....(((((((((((((((-((........))))))))....)))))))))...))).)).))))) ( -28.80) >DroEre_CAF1 41266 88 - 1 -UUGUGGCACGGCGCAG-------------------CGGAAACGGAAACUAUUUGCUGCCAUU-UGGGCAAUGGCAAACGGCAA-UGGCAAGUGGCAGAAUGCGAGCAUU -.....((....((((.-------------------((....))...........((((((((-(.(.((.((.(....).)).-)).))))))))))..)))).))... ( -28.60) >DroYak_CAF1 43414 91 - 1 GGGGCGGCAGGGAGAAG-------------------CGGAAACGGAAACUAUUUGCUGCCAUUUUGGGCAAUGGCAAGUGGCAAGUGGCAAAUGGCAGAAUGCGAGCAUU ..(((((((((.((...-------------------((....))....)).))))))))).......(((.((.((..((.(....).))..)).))...)))....... ( -27.80) >consensus AGGGGUGUGUGGAUUGG___________________CGGAAACGGAAACUAUUUGCUGCCAUU_UGGGCAAUGGCAAAUGGCAAAUAGCAAAUGGCAGAAUGCGAGCAUU ......((((..........................((....))....(((((((((((((((....((....)).))))))....)))))))))....))))....... (-22.96 = -22.52 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:49 2006