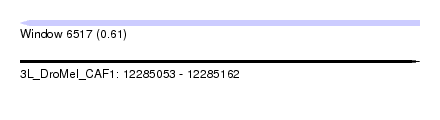
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,285,053 – 12,285,162 |
| Length | 109 |
| Max. P | 0.613566 |

| Location | 12,285,053 – 12,285,162 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 88.30 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -24.08 |
| Energy contribution | -23.47 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
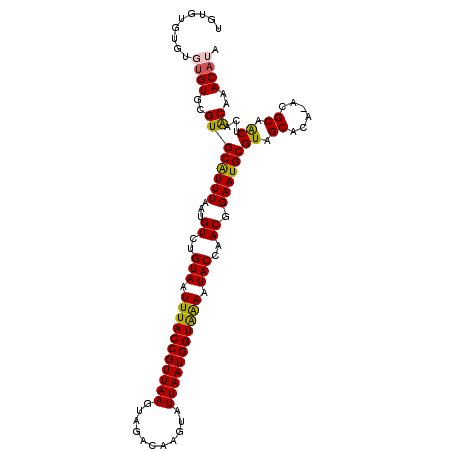
>3L_DroMel_CAF1 12285053 109 - 23771897 AGUGUGCGUGUGUGCGUGCGUUUAAUGUCUGUAAUUUACCGUUAAGUAGACAAGUAUUAAUGGUAGAAUACCAACGGAAUGCGUAGCAGA-ACGCAGCUCCACAAACAUA .(((.((((((.(((.(((((....(((..(((.(((((((((((...........))))))))))).)))..)))....))))))))..-)))).))..)))....... ( -33.50) >DroVir_CAF1 22759 101 - 1 UG--------UGUGCGUGCAUUUAAUGUCUGUAAUUUACCGUUAAGUAGACAAGUAUUAAUGGUGAAAUACCAACGGAAUGCGUAGCGCU-GCGCAACUUAACAAACAUA ((--------(((((((((((((...((..(((.(((((((((((...........))))))))))).)))..)).))))))...)))).-))))).............. ( -29.00) >DroGri_CAF1 21934 101 - 1 UG--------UGUGCGUGCAUUUAAUGUCUGUAAUUUACCGUUAAGUAGACAAGUAUUAAUGGUGAAAUACCAACGGAAUGCGUAGCGCU-GCGCAACUUAACAAACAUA ((--------(((((((((((((...((..(((.(((((((((((...........))))))))))).)))..)).))))))...)))).-))))).............. ( -29.00) >DroMoj_CAF1 22788 109 - 1 UGUGUAUGUGUGUGUGUGCAUUUAAUGUCUGUAAUUUACCGUUAAGUAGACAAGUAUUAAUGGUGAAAUACCAACGGAAUGCGUAGCGCU-GCGCAACUUAACAAACAUA ((((((.((((.....(((((((...((..(((.(((((((((((...........))))))))))).)))..)).)))))))..)))))-))))).............. ( -29.90) >DroAna_CAF1 17044 108 - 1 -GGGUGUGUGGGUGCGUGCGUUUAAUGUCUGUAAUUUACCGUUAAGUAGACAAGUAUUAAUGGUAGAAUACCAACGGAAUGCGUAGCAGA-ACGCAGCUCCGCAAACAUA -..((.((((((.((.(((((((..(((..(((.(((((((((((...........))))))))))).)))..)))...(((...)))))-))))))))))))).))... ( -35.10) >DroPer_CAF1 24372 110 - 1 UGUGUGUGUGUGUGCGUGCGUUUAAUGUCUGUAAUUUACCGUUAAGUAGACAAGUAUUAAUGGUAAAAUACCAACGGAAUGCGUAGCAGAAACGCAGCUCCACAAACAUA (((((.((((.(.((.(((((((..(((..(((.(((((((((((...........))))))))))).)))..)))...(((...))).))))))))).))))).))))) ( -34.10) >consensus UGUGUGUGUGUGUGCGUGCAUUUAAUGUCUGUAAUUUACCGUUAAGUAGACAAGUAUUAAUGGUAAAAUACCAACGGAAUGCGUAGCACA_ACGCAACUCAACAAACAUA .........((((..((((((((...((..(((.(((((((((((...........))))))))))).)))..)).))))))((.((......)).))...))..)))). (-24.08 = -23.47 + -0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:45 2006