
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,283,586 – 12,283,693 |
| Length | 107 |
| Max. P | 0.958242 |

| Location | 12,283,586 – 12,283,693 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.64 |
| Mean single sequence MFE | -19.05 |
| Consensus MFE | -12.55 |
| Energy contribution | -13.13 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12283586 107 - 23771897 CCAUUCCCAUCCCCUUUUCCCAUUCCAUU------CUUGGCCAUAUCCUUAUACCCAUUCUCAUC--CAUAUCCUUAGUGGAUGUGACUAGACUAAGAUGGAUUAAGCUCCAAGU .......................(((((.------(((((.((((....)))......((.((((--(((.......))))))).))...).))))))))))............. ( -19.10) >DroPse_CAF1 21732 90 - 1 UCAUCCUCAUCCACAU------CCACAU------------CCACAUCCA------CGUCGUCAUCAUCAUGGUCGUACUGGAUGUGACUAGACUAAGAUGGAUUAAGC-CCAAGU ..............((------(((...------------.((((((((------.((((((((....)))).)).)))))))))).((......)).))))).....-...... ( -19.40) >DroSec_CAF1 18635 113 - 1 CCAUUCCCAAUCCCACUUCCCAUCCCAUCCUUAUCCUUGGCCAUAUCCUUAUACCCAUUCUCAUC--CAUAUCCUUAGUGGAUGUGACUAGACUAAGAUGGAUUAAGCUCCAAGU ........................(((((.(((((..(((..(((....)))..))).((.((((--(((.......))))))).))...)).)))))))).............. ( -18.50) >DroSim_CAF1 18929 113 - 1 CCAUUCCAAAUCCCACUUCCCAUCCCAUCCUUAUCCUUGGCCAUAUCCUUAUACCCAUUCUCAUC--CAUAUCCUUAGUGGAUGUGACUAGACUAAGAUGGAUUAAGCUCCAAGU ........................(((((.(((((..(((..(((....)))..))).((.((((--(((.......))))))).))...)).)))))))).............. ( -18.50) >DroEre_CAF1 18678 107 - 1 CCAUCCCCAUCCCCAU------UCCCAUCCUUAUCCUUGGCCAUAUCCUUAUACCCAUGCUCAUC--CGUAUCCUUAGUGGAUGUGACUAGACUAAGAUGGAUUAAGCCCCAAGU ................------..(((((.(((...((((.(((((((...((...((((.....--.))))...))..))))))).))))..)))))))).............. ( -19.00) >DroYak_CAF1 19320 101 - 1 CCAUC------CCCAU------UCUCAUCCUUAUCCUUGGCCAUAUCCUCGCACCCAUUCUCAUC--CAUAUCCUUAGUGGAUGUGACUAGACUAAGAUGGAUUAAGCCCCAAGU .....------.....------.............(((((....((((((........((.((((--(((.......))))))).)).........)).))))......))))). ( -19.83) >consensus CCAUCCCCAUCCCCAU______UCCCAUCCUUAUCCUUGGCCAUAUCCUUAUACCCAUUCUCAUC__CAUAUCCUUAGUGGAUGUGACUAGACUAAGAUGGAUUAAGCUCCAAGU ...................................(((((....((((...................(((((((.....))))))).((......))..))))......))))). (-12.55 = -13.13 + 0.59)

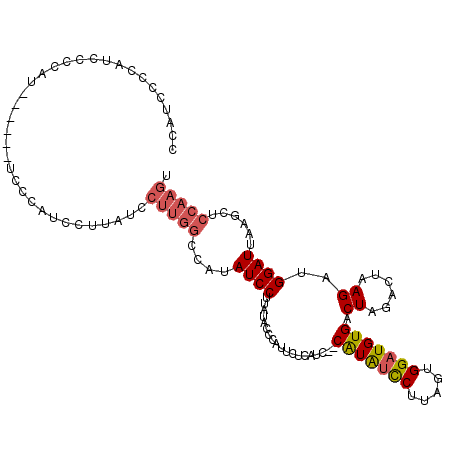
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:44 2006