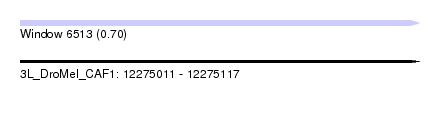
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,275,011 – 12,275,117 |
| Length | 106 |
| Max. P | 0.695824 |

| Location | 12,275,011 – 12,275,117 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.42 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -18.41 |
| Energy contribution | -18.38 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12275011 106 + 23771897 -----ACUGC---------UAGCUACUGUUUGCAAAUGGAUCAAGGAUAUUUACCAGAGGCACGGGCUAGAUAUUCCUGAGAAAUGUAAACAGCGCCAAAUGCAAACAGUUCCAAUCAAA -----.....---------..(..((((((((((..(((.(((.(((((((((((.(.....).)).))))))))).))).....((.....)).)))..))))))))))..)....... ( -27.60) >DroVir_CAF1 12653 105 + 1 -----UCU-------G---UUGCUUCUGUUUGCAAAUGGAUCAAGGAUAUUUACCGAUGGCACGGGCUAGAUAUUCCUGAGAUAUGUAAACAGCACUGAAUGCAAACAGUUCAAAUCAAA -----.((-------(---((((..(((((((((..((..(((.((((((((((((......)))..))))))))).)))..)))))))))))........)).)))))........... ( -26.60) >DroGri_CAF1 12098 97 + 1 -----------------------UGCUGUUUGCAAAUGGAUCAAGGAUAUUUACCGGAGGCACUGGCUAGAUACUCCUGAGAUAAGUAAACAGCACAGAAUGCAAACAGUUCAAAUCAAA -----------------------((((((((((...((..(((.(((((((((((((.....)))).)))))).))))))..)).))))))))))..((((.......))))........ ( -28.10) >DroWil_CAF1 16686 111 + 1 UUGCUACUAC---------AACUUGAUGUUUGCAAAUGGAUCAAGGAUAUUUACCGUAGGGACGGGCUAGAUAUUCCUGAGAAAUGUAAACAGCGCAAAACGCAAACUGUUCAAAUCAAA ..........---------...((((((((((((......(((.(((((((((((((....))))..))))))))).)))....))))))).(((.....)))...........))))). ( -26.00) >DroMoj_CAF1 12875 105 + 1 -----UCU-------G---UUGCUGUUGUUUGCCAAUGGAUCAAGGAUAUUUACCGAAGGCACGGGCUAGAUAUUCCUGAGAUAUGUAAACAGUACUGAAUGCAAACAGUUCAAAUCAAA -----..(-------(---..((((((((((((..(((..(((.((((((((((((......)))..))))))))).)))..))))))))).(((.....))).)))))).))....... ( -24.80) >DroAna_CAF1 8356 115 + 1 -----ACGGCUUCUGGCCGUUGCCACUGUUUGCAAAUGGAUCAAGGAUAUUUACCAGAGGCACGGGCUAGAUAUUCCUGAGAAAUGUAAACAGCGCAAAAUGCAAACAGUUCCAAUCAAA -----(((((.....)))))((..((((((((((......(((.(((((((((((.(.....).)).))))))))).)))(...((....))...)....))))))))))..))...... ( -31.90) >consensus _____ACU___________UUGCUGCUGUUUGCAAAUGGAUCAAGGAUAUUUACCGGAGGCACGGGCUAGAUAUUCCUGAGAAAUGUAAACAGCACAAAAUGCAAACAGUUCAAAUCAAA ..........................((((((((......(((.(((((((((((.........)).))))))))).)))....))))))))(((.....)))................. (-18.41 = -18.38 + -0.03)

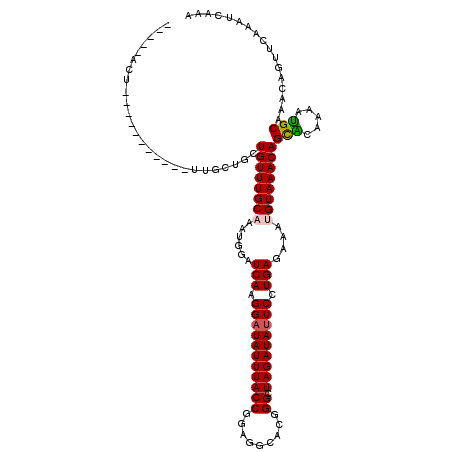

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:41 2006