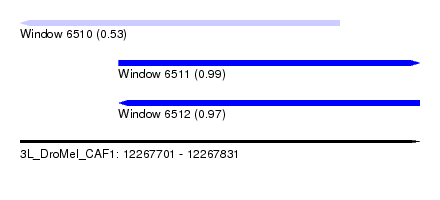
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,267,701 – 12,267,831 |
| Length | 130 |
| Max. P | 0.993156 |

| Location | 12,267,701 – 12,267,805 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 82.38 |
| Mean single sequence MFE | -25.01 |
| Consensus MFE | -15.26 |
| Energy contribution | -15.22 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.533886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12267701 104 - 23771897 GGCAAUUAAAUGCAGUCCAAUUAAAUGUACUGAGAGCUGCAAUCGACUAUUUUCGCAUUUAU-------UUGUCAUUUGGCAGAACAAACUGAGUGCAAGCUGGUCUAGAG ..........(((((..(...............)..)))))...(((((.(((.((((((((-------(((((....))))))......)))))))))).)))))..... ( -22.76) >DroSec_CAF1 2736 104 - 1 GGCAAUUAAAUGCAGUCCAAUUAAAUGUACUGAGAGCUGCAAUCGACUAUUUUCGCAUUUAU-------UUGCCAUGUGGCAGAACAAACUGAGUGCAAGCUGUUCUAAAG ............((((.((......)).))))(((((.((....((......))((((((((-------(((((....))))))......)))))))..)).))))).... ( -24.60) >DroSim_CAF1 2685 89 - 1 GGCAAUUAAAUGCAGUCCAAUUAAAUGUACUG---------------UAUUUUCGCAUUUAU-------UUGCCAUUGGGCAGAACAAACUGAGUGCAAGCUGUUCUAAAG (((....(((((((((.((......)).))))---------------)))))..((((((((-------(((((....))))))......)))))))..)))......... ( -24.40) >DroEre_CAF1 2779 103 - 1 GGCAAUUAAAUGCAGUCCAAUUAAAUGCACUCACAACGGCAAUUGACUAUUUUCGCAUUUAU-------UUGCCAUUCGGCUGAACGAGACAAGUGCAAGCUGA-CUAAAG (((((.(((((((((((.(((....(((.(.......)))))))))))......))))))).-------)))))..((((((..((.......))...))))))-...... ( -26.50) >DroYak_CAF1 2890 110 - 1 GGCAAUUAAAUGCAGUCCAAUUAAAUGCACUCAGAACUGCAAUCGACUAAUUUCGCAUUUAUUUACGAUUUGCCAUUUGGCUGAAUAAAAUAAGUGCAAGCUGA-GUAAAG .(((.((((.((.....)).)))).)))((((((..........((......))((((((((((...(((((((....))).)))).))))))))))...))))-)).... ( -26.80) >consensus GGCAAUUAAAUGCAGUCCAAUUAAAUGUACUGAGAACUGCAAUCGACUAUUUUCGCAUUUAU_______UUGCCAUUUGGCAGAACAAACUGAGUGCAAGCUGAUCUAAAG (((.......((((...........)))).........................(((((((........(((((....))))).......)))))))..)))......... (-15.26 = -15.22 + -0.04)

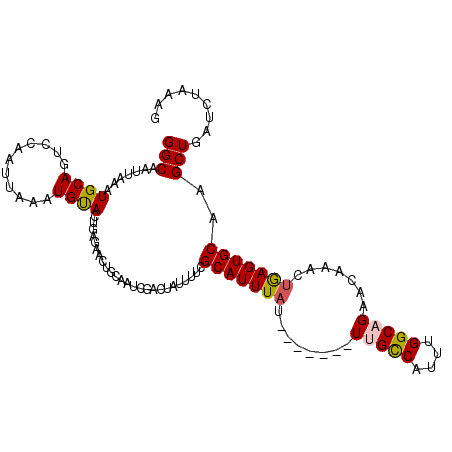
| Location | 12,267,733 – 12,267,831 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 82.09 |
| Mean single sequence MFE | -24.07 |
| Consensus MFE | -15.98 |
| Energy contribution | -16.86 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12267733 98 + 23771897 CAAAUGACAA-------AUAAAUGCGAAAAUAGUCGAUUGCAGCUCUCAGUACAUUUAAUUGGACUGCAUUUAAUUGCCAUUUGGAUAUAGAAAAACUAUGCAGA- ((((((.(((-------.(((((.(((......)))...((((.((.((((.......))))))))))))))).))).))))))(.(((((.....))))))...- ( -20.10) >DroSec_CAF1 2768 99 + 1 CACAUGGCAA-------AUAAAUGCGAAAAUAGUCGAUUGCAGCUCUCAGUACAUUUAAUUGGACUGCAUUUAAUUGCCAUUUGGAAAUGGAAAAACUAUGCAGAA ((.(((((((-------.(((((.(((......)))...((((.((.((((.......))))))))))))))).))))))).))...((((.....))))...... ( -22.50) >DroSim_CAF1 2717 84 + 1 CCAAUGGCAA-------AUAAAUGCGAAAAUA---------------CAGUACAUUUAAUUGGACUGCAUUUAAUUGCCAUUUGGAUGCAGAAAAACUAUGCUAAA ((((((((((-------.((((((((......---------------((((.......))))...)))))))).)))))).))))..(((.........))).... ( -22.50) >DroEre_CAF1 2810 97 + 1 CGAAUGGCAA-------AUAAAUGCGAAAAUAGUCAAUUGCCGUUGUGAGUGCAUUUAAUUGGACUGCAUUUAAUUGCCAUUUGAAUAUAGAAAAAGUAUGCA--A ((((((((((-------.((((((((....((((.((.((((.......).))).)).))))...)))))))).))))))))))...................--. ( -26.30) >DroYak_CAF1 2921 104 + 1 CAAAUGGCAAAUCGUAAAUAAAUGCGAAAUUAGUCGAUUGCAGUUCUGAGUGCAUUUAAUUGGACUGCAUUUAAUUGCCAUUUGAAUGUAGAAAAACUAUGCA--A ((((((((((.(((((......)))))........((.((((((((...............)))))))).))..))))))))))..((((.........))))--. ( -28.96) >consensus CAAAUGGCAA_______AUAAAUGCGAAAAUAGUCGAUUGCAGCUCUCAGUACAUUUAAUUGGACUGCAUUUAAUUGCCAUUUGGAUAUAGAAAAACUAUGCA_AA ((((((((((........(((((((((......)).............(((.((......)).)))))))))).))))))))))...................... (-15.98 = -16.86 + 0.88)

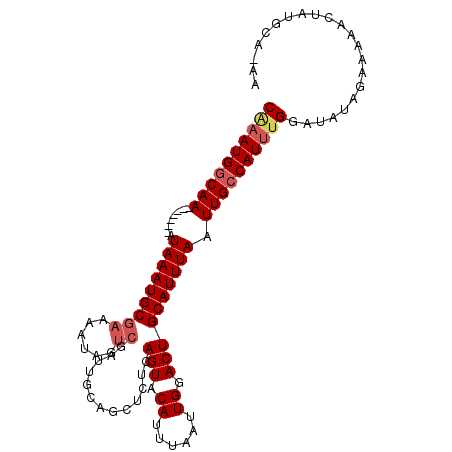
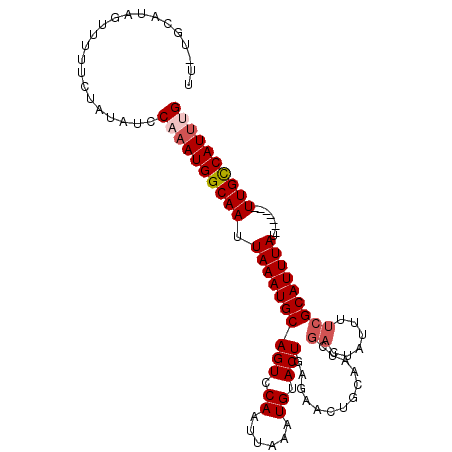
| Location | 12,267,733 – 12,267,831 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 82.09 |
| Mean single sequence MFE | -21.30 |
| Consensus MFE | -14.36 |
| Energy contribution | -15.20 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12267733 98 - 23771897 -UCUGCAUAGUUUUUCUAUAUCCAAAUGGCAAUUAAAUGCAGUCCAAUUAAAUGUACUGAGAGCUGCAAUCGACUAUUUUCGCAUUUAU-------UUGUCAUUUG -.....((((.....))))...((((((((((.((((((((((.((......)).)))(((((..((....).)..)))))))))))).-------)))))))))) ( -23.10) >DroSec_CAF1 2768 99 - 1 UUCUGCAUAGUUUUUCCAUUUCCAAAUGGCAAUUAAAUGCAGUCCAAUUAAAUGUACUGAGAGCUGCAAUCGACUAUUUUCGCAUUUAU-------UUGCCAUGUG ......................((.(((((((.((((((((((.((......)).)))(((((..((....).)..)))))))))))).-------))))))).)) ( -21.40) >DroSim_CAF1 2717 84 - 1 UUUAGCAUAGUUUUUCUGCAUCCAAAUGGCAAUUAAAUGCAGUCCAAUUAAAUGUACUG---------------UAUUUUCGCAUUUAU-------UUGCCAUUGG ....(((.........)))..((((.((((((.((((((((((.((......)).)))(---------------......)))))))).-------)))))))))) ( -21.20) >DroEre_CAF1 2810 97 - 1 U--UGCAUACUUUUUCUAUAUUCAAAUGGCAAUUAAAUGCAGUCCAAUUAAAUGCACUCACAACGGCAAUUGACUAUUUUCGCAUUUAU-------UUGCCAUUCG .--.....................((((((((.(((((((((((.(((....(((.(.......)))))))))))......))))))).-------)))))))).. ( -20.20) >DroYak_CAF1 2921 104 - 1 U--UGCAUAGUUUUUCUACAUUCAAAUGGCAAUUAAAUGCAGUCCAAUUAAAUGCACUCAGAACUGCAAUCGACUAAUUUCGCAUUUAUUUACGAUUUGCCAUUUG .--...................((((((((((.(((((((............((((........))))...((......)))))))))........)))))))))) ( -20.60) >consensus UU_UGCAUAGUUUUUCUAUAUCCAAAUGGCAAUUAAAUGCAGUCCAAUUAAAUGUACUGAGAACUGCAAUCGACUAUUUUCGCAUUUAU_______UUGCCAUUUG ......................((((((((((.((((((((((.((......)).))).............((......)))))))))........)))))))))) (-14.36 = -15.20 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:40 2006