
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,262,176 – 12,262,298 |
| Length | 122 |
| Max. P | 0.962033 |

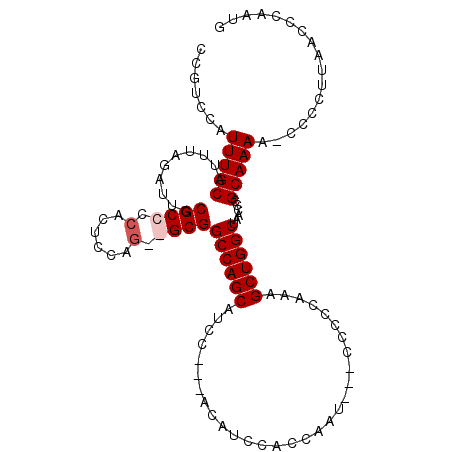
| Location | 12,262,176 – 12,262,271 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 86.34 |
| Mean single sequence MFE | -21.82 |
| Consensus MFE | -17.74 |
| Energy contribution | -18.48 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12262176 95 + 23771897 CCGUCCAUUUGCAUUUAGAUUGCGCCCCACUCCAG--GCGGCCAGCAU---------CCACGAAU---CUCCCAAAGCUGGCUAACCCGCAAAA-CCCCUUAACCCAAUG .......(((((..........((((........)--)))((((((..---------........---........))))))......))))).-............... ( -21.17) >DroSec_CAF1 15669 103 + 1 CCGUCCAUUUGCAUUUAGAUUGCGCCCCACUCCAGGGGCGGCCAGCAUAC---ACAUCCACCAAU---CCCCCAAAGCUGGCUAACCCGCAAAA-CCCCUUAACCCAAUG .......(((((..........((((((......))))))((((((....---............---........))))))......))))).-............... ( -28.16) >DroSim_CAF1 18107 103 + 1 CCGUCCAUUUGCAUUUAGAUUGCGCCCCACUCCAGUGGCGGCCAGCAUCC---ACAUCCACCAAU---CCCCCAAAGCUGGCUAACCCGCAAAA-CCCCUUAACCCAAUG .......(((((..........((((.(......).))))((((((....---............---........))))))......))))).-............... ( -21.06) >DroYak_CAF1 16599 108 + 1 UCGUCCAUUUGCAUUUAGAUUGCGCCCCACUCCAA--GCGGCCAGCAUCCUCCACUCCCCUCAAAUACCCCCAAAAGCUGGCGAACCCGCAAAAGCCCCCAAACCCAAUG .......(((((..........(((..........--)))((((((..............................))))))......)))))................. ( -16.91) >consensus CCGUCCAUUUGCAUUUAGAUUGCGCCCCACUCCAG__GCGGCCAGCAUCC___ACAUCCACCAAU___CCCCCAAAGCUGGCUAACCCGCAAAA_CCCCUUAACCCAAUG .......(((((..........((((.(......).))))((((((..............................))))))......)))))................. (-17.74 = -18.48 + 0.75)


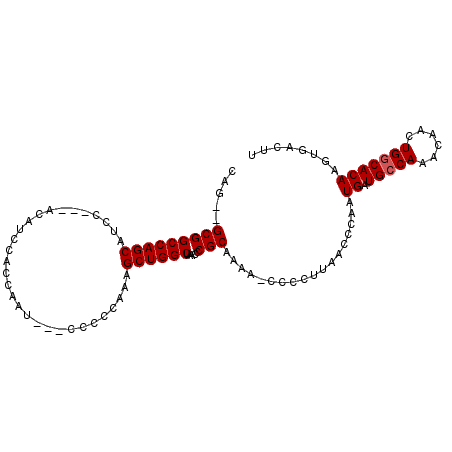
| Location | 12,262,208 – 12,262,298 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 85.17 |
| Mean single sequence MFE | -20.43 |
| Consensus MFE | -16.78 |
| Energy contribution | -17.04 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.962033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12262208 90 + 23771897 CAG--GCGGCCAGCAU---------CCACGAAU---CUCCCAAAGCUGGCUAACCCGCAAAA-CCCCUUAACCCAAUGAUGCCAAACAACUGGCACAAGUGACUU ..(--(.(((((((..---------........---........)))))))...))......-.............((.(((((......)))))))........ ( -19.67) >DroSec_CAF1 15701 98 + 1 CAGGGGCGGCCAGCAUAC---ACAUCCACCAAU---CCCCCAAAGCUGGCUAACCCGCAAAA-CCCCUUAACCCAAUGAUGCCAAACAACUGGCACAAGUGAUUU ..(((..(((((((....---............---........)))))))..)))......-.............((.(((((......)))))))........ ( -25.96) >DroSim_CAF1 18139 98 + 1 CAGUGGCGGCCAGCAUCC---ACAUCCACCAAU---CCCCCAAAGCUGGCUAACCCGCAAAA-CCCCUUAACCCAAUGAUGCCAAACAACUGGCACAAGUGACUU ..((((.(((((((....---............---........)))))))...))))....-.............((.(((((......)))))))........ ( -23.26) >DroYak_CAF1 16631 103 + 1 CAA--GCGGCCAGCAUCCUCCACUCCCCUCAAAUACCCCCAAAAGCUGGCGAACCCGCAAAAGCCCCCAAACCCAAUGAUGCCAAACAACUGCCACAAGUGACUU ...--(((((((((..............................))))))......((....))...............)))......(((......)))..... ( -12.81) >consensus CAG__GCGGCCAGCAUCC___ACAUCCACCAAU___CCCCCAAAGCUGGCUAACCCGCAAAA_CCCCUUAACCCAAUGAUGCCAAACAACUGGCACAAGUGACUU .....(((((((((..............................)))))).....)))..................((.(((((......)))))))........ (-16.78 = -17.04 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:34 2006