| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,258,656 – 12,258,760 |
| Length | 104 |
| Max. P | 0.500000 |

| Location | 12,258,656 – 12,258,760 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
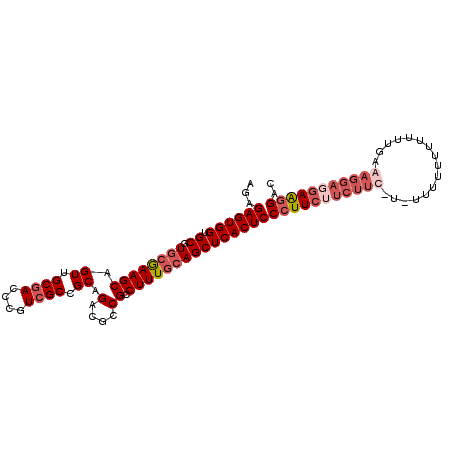
| Reading direction | forward |
| Mean pairwise identity | 86.61 |
| Mean single sequence MFE | -30.99 |
| Consensus MFE | -26.31 |
| Energy contribution | -27.62 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.85 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12258656 104 + 23771897 GUCCCUCCUCCUUUCAAAAAAAAAAAAAAAAAGAAGAAGGGAGUGAGCUGCAAAGGCGGAGUCUGCGGCGACGGGUCGCAACUGCUUCGCACGCACCACUCUCU ....(((((((((((....................)))))))).))).(((....(((((((.((((((.....))))))...)))))))..)))......... ( -33.45) >DroSec_CAF1 12138 102 + 1 GUCCUUCCUCCUUUCCAAAAAAAAAA-A-GAAGAUGAAGGGAGUGAGCUGCAAAGGCGGCGUCUGCGGCUACGGGUCGCAACUGCUUCGCACGCACCACUCUCU ..(((((.((((((..........))-)-)..)).)))))(((((.(((((..((((((....((((((.....)))))).)))))).))).))..)))))... ( -31.80) >DroSim_CAF1 14592 103 + 1 GUCCUUCCUCCUUUCAAAAAAAAAAA-AUAAAGAAGAAGGGAGUGAGCUGCAAAGGCGGCGUCUGCGGCGACGGGUCGCAACUGCUUCGCACGCACCACUCUCU ..(((((.((................-.....)).)))))(((((.(((((....)))))...(((((((((((.......)))..)))).)))).)))))... ( -32.30) >DroYak_CAF1 13077 89 + 1 GUCCUUCCCCCGU------AAAAAAA-A-GA-----AAUGGAGUGAGCUCGAAAGGCGGAGUCUGCGGCGACGGGUCGCAACUGCUUUGCACGCACCACUC--U .............------.......-.-..-----...((((((.((.(....)(((((((.((((((.....))))))...)))))))..))..)))))--) ( -26.40) >consensus GUCCUUCCUCCUUUCAAAAAAAAAAA_A_AAAGAAGAAGGGAGUGAGCUGCAAAGGCGGAGUCUGCGGCGACGGGUCGCAACUGCUUCGCACGCACCACUCUCU .....................................((((((((.(.(((....(((((((.((((((.....))))))...)))))))..)))))))))))) (-26.31 = -27.62 + 1.31)

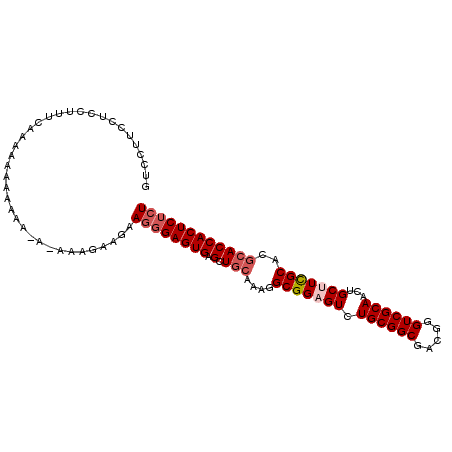
| Location | 12,258,656 – 12,258,760 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 86.61 |
| Mean single sequence MFE | -34.24 |
| Consensus MFE | -25.64 |
| Energy contribution | -28.51 |
| Covariance contribution | 2.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12258656 104 - 23771897 AGAGAGUGGUGCGUGCGAAGCAGUUGCGACCCGUCGCCGCAGACUCCGCCUUUGCAGCUCACUCCCUUCUUCUUUUUUUUUUUUUUUUUGAAAGGAGGAGGGAC ...((((((.((.((((((((.((.((((....)))).)).(....)).)))))))))))))))(((((((((((((............))))))))))))).. ( -39.50) >DroSec_CAF1 12138 102 - 1 AGAGAGUGGUGCGUGCGAAGCAGUUGCGACCCGUAGCCGCAGACGCCGCCUUUGCAGCUCACUCCCUUCAUCUUC-U-UUUUUUUUUUGGAAAGGAGGAAGGAC ...((((((.((.((((((((.((((((...)))))).((....)).).)))))))))))))))(((((..((((-(-((((......)))))))))))))).. ( -37.50) >DroSim_CAF1 14592 103 - 1 AGAGAGUGGUGCGUGCGAAGCAGUUGCGACCCGUCGCCGCAGACGCCGCCUUUGCAGCUCACUCCCUUCUUCUUUAU-UUUUUUUUUUUGAAAGGAGGAAGGAC ...((((((.((.(((((((.....(((...((((......)))).))))))))))))))))))(((((((((((..-.............))))))))))).. ( -37.76) >DroYak_CAF1 13077 89 - 1 A--GAGUGGUGCGUGCAAAGCAGUUGCGACCCGUCGCCGCAGACUCCGCCUUUCGAGCUCACUCCAUU-----UC-U-UUUUUUU------ACGGGGGAAGGAC .--((((((((((.((...((....))((....))))))))(.(((........))))))))))....-----((-(-((((((.------...))))))))). ( -22.20) >consensus AGAGAGUGGUGCGUGCGAAGCAGUUGCGACCCGUCGCCGCAGACGCCGCCUUUGCAGCUCACUCCCUUCUUCUUC_U_UUUUUUUUUUUGAAAGGAGGAAGGAC ...((((((.((.((((((((.((.((((....)))).)).(....)).)))))))))))))))(((((((((((................))))))))))).. (-25.64 = -28.51 + 2.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:33 2006