
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,247,952 – 12,248,060 |
| Length | 108 |
| Max. P | 0.993537 |

| Location | 12,247,952 – 12,248,060 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.64 |
| Mean single sequence MFE | -39.42 |
| Consensus MFE | -11.48 |
| Energy contribution | -11.45 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.29 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.735285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12247952 108 + 23771897 ACGAUGGGAAGUACCUGGAGACAUCGCCGCGCUUCAGCGUCGGCAGCUUCUGGGCCUUGAUGGCCUUCAGUUUGAUUGCCAGGACAUUCUCCAGGCUCUUACCG---------CUCU--- ..(((((.(((..((((((((.....((((((....)))).((((((..(((((((.....))))..)))...).))))).))....))))))))..))).)))---------.)).--- ( -45.00) >DroPse_CAF1 1253 120 + 1 ACGAUGGAAAAUACCUAGGGACAUCACCGCGCCUCAGGGCGGGCAGCUUUUGUCCCCUAAUGGCCUUCAGUUUCACGGCCAAGGCAUUUUCCAAGCUCUUGCCAUGCACCAAGCCCUAGU ....((((((((.(((.((((((...((.((((....)))))).......))))))....(((((...........)))))))).)))))))).(((..(((...)))...)))...... ( -40.40) >DroWil_CAF1 3523 108 + 1 AUGAUGGAAAAUACCUAGCGACAUCACCGCGCUUCAAGGCAGGUAGCUUCUGGCCUCUAAGUGCUUUCAAUUUCAAUGCUAGGGAAUUUUCCAGACUUUUGCCG---------AUCU--- ....((((((((.(((((((....)...((((((..(((((((.....))).))))..)))))).............))))))..))))))))...........---------....--- ( -34.20) >DroMoj_CAF1 1266 108 + 1 AGGAUGGAUAAUACCUAGCGACAUCGCCGCGCUUCAAUGUUGGUAUCUUUUGACCCGAAAGGGCUUGCAAACUUUUUGCUAGGGCAUUUUCCAAGCGUUUGCCC---------UUCU--- (((.((((.(((.((((((((....((((.((......))))))....((((.(((....)))....))))....))))))))..))).)))).((....))))---------)...--- ( -31.30) >DroAna_CAF1 1438 108 + 1 AGGAGGGGAAGUACCUGGCGACAUCGCCGCGCUUCAGCGCCGGCAGCUUCUGGCCCUUGACAGCCUUCAACUUGAGGGCCAGGACCUUCUCCAGUCUCAUAGCG---------CCCU--- (((.(((((((..((.((((....))))((((....)))).))....((((((((((..(...........)..)))))))))).))))))).((......)).---------.)))--- ( -45.20) >DroPer_CAF1 1253 120 + 1 ACGAUGGAAAAUACCUAGGGACAUCACCGCGCCUCAGGGCGGGCAGCUUUUGUCCCCUAAUGGCCUUCAGUUUCACGGCCAAGGCAUUUUCCAAGCUCUUGCCAUGCACCAAGCCCUAGU ....((((((((.(((.((((((...((.((((....)))))).......))))))....(((((...........)))))))).)))))))).(((..(((...)))...)))...... ( -40.40) >consensus ACGAUGGAAAAUACCUAGCGACAUCACCGCGCUUCAGGGCCGGCAGCUUCUGGCCCCUAAUGGCCUUCAAUUUCACGGCCAGGGCAUUUUCCAAGCUCUUGCCG_________CCCU___ ....((((.(((.(((............(((((........))).))..............(((.............))).))).))).))))........................... (-11.48 = -11.45 + -0.03)



| Location | 12,247,952 – 12,248,060 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.64 |
| Mean single sequence MFE | -43.78 |
| Consensus MFE | -18.17 |
| Energy contribution | -19.65 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.42 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
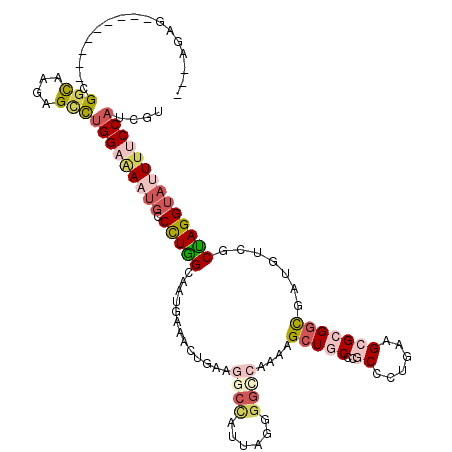
>3L_DroMel_CAF1 12247952 108 - 23771897 ---AGAG---------CGGUAAGAGCCUGGAGAAUGUCCUGGCAAUCAAACUGAAGGCCAUCAAGGCCCAGAAGCUGCCGACGCUGAAGCGCGGCGAUGUCUCCAGGUACUUCCCAUCGU ---.((.---------.((.(((.(((((((((.(((....)))(((...(((..((((.....))))))).....((((.(((....)))))))))).))))))))).))).)).)).. ( -47.00) >DroPse_CAF1 1253 120 - 1 ACUAGGGCUUGGUGCAUGGCAAGAGCUUGGAAAAUGCCUUGGCCGUGAAACUGAAGGCCAUUAGGGGACAAAAGCUGCCCGCCCUGAGGCGCGGUGAUGUCCCUAGGUAUUUUCCAUCGU ....((((((..(((...))).))))))((((((((((((((((...........)))))...(((((((...(((((..(((....))))))))..))))))))))))))))))..... ( -53.10) >DroWil_CAF1 3523 108 - 1 ---AGAU---------CGGCAAAAGUCUGGAAAAUUCCCUAGCAUUGAAAUUGAAAGCACUUAGAGGCCAGAAGCUACCUGCCUUGAAGCGCGGUGAUGUCGCUAGGUAUUUUCCAUCAU ---.(((---------.(((....))).(((((((..((((((...((.((((...((.(((.(((((.((.......))))))).))).))..)))).)))))))).)))))))))).. ( -33.00) >DroMoj_CAF1 1266 108 - 1 ---AGAA---------GGGCAAACGCUUGGAAAAUGCCCUAGCAAAAAGUUUGCAAGCCCUUUCGGGUCAAAAGAUACCAACAUUGAAGCGCGGCGAUGUCGCUAGGUAUUAUCCAUCCU ---...(---------((((....)).((((.((((((.((((.....((((....((((....))))....))))....((((((........)))))).)))))))))).)))).))) ( -30.00) >DroAna_CAF1 1438 108 - 1 ---AGGG---------CGCUAUGAGACUGGAGAAGGUCCUGGCCCUCAAGUUGAAGGCUGUCAAGGGCCAGAAGCUGCCGGCGCUGAAGCGCGGCGAUGUCGCCAGGUACUUCCCCUCCU ---((((---------.(((....((((......))))((((((((..(((.....)))....)))))))).)))((((.((((....))))((((....)))).))))....))))... ( -46.50) >DroPer_CAF1 1253 120 - 1 ACUAGGGCUUGGUGCAUGGCAAGAGCUUGGAAAAUGCCUUGGCCGUGAAACUGAAGGCCAUUAGGGGACAAAAGCUGCCCGCCCUGAGGCGCGGUGAUGUCCCUAGGUAUUUUCCAUCGU ....((((((..(((...))).))))))((((((((((((((((...........)))))...(((((((...(((((..(((....))))))))..))))))))))))))))))..... ( -53.10) >consensus ___AGAG_________CGGCAAGAGCCUGGAAAAUGCCCUGGCAAUGAAACUGAAGGCCAUUAGGGGCCAAAAGCUGCCCGCCCUGAAGCGCGGCGAUGUCGCUAGGUAUUUUCCAUCGU .................(((....)))(((((((((.(((((.............((((......))))....(((((..((......))))))).......)))))))))))))).... (-18.17 = -19.65 + 1.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:26 2006