| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,215,447 – 12,215,538 |
| Length | 91 |
| Max. P | 0.878055 |

| Location | 12,215,447 – 12,215,538 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 81.27 |
| Mean single sequence MFE | -17.67 |
| Consensus MFE | -14.10 |
| Energy contribution | -13.97 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
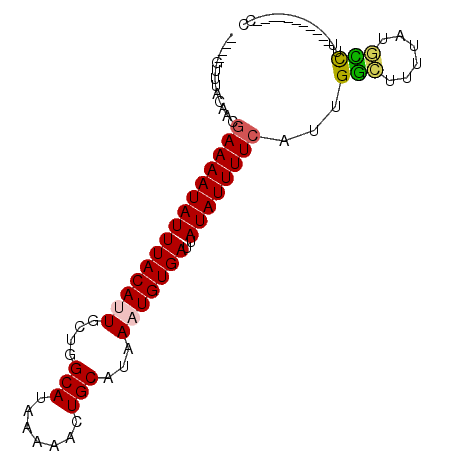
>3L_DroMel_CAF1 12215447 91 + 23771897 ----GUUUACUACGAAAAUAUUUACAUUGCUGGCAUAAAAACUGCAUAAAUGUGAUUAUAUUUUCAUUGGCCUUUAUGCCUUAUGCCGUCGUUUU ----....((.(((((((((((((((((....(((.......)))...)))))))..))))))))...(((......))).......)).))... ( -17.90) >DroVir_CAF1 32437 80 + 1 ----GUUUACAGCGAAAAUAUUUACACUGCAGGCAUAAAAACUGCAUAAAUGUGAUUAUAUUUUCAUUGGCUUUUAUGUUGG-----------CC ----.........(((((((((((((.(((((.........)))))....)))))..))))))))...((((........))-----------)) ( -18.40) >DroWil_CAF1 34453 87 + 1 ----GUUUACAACGAAAAUAUUUACAUUGCUGGCAUAAAAACUGCAUAAAUGUGAUUAUAUUUUCAUUGAGUUUUAUUUUUCAUACUCA----UC ----.........(((((((((((((((....(((.......)))...)))))))..))))))))..(((((............)))))----.. ( -17.30) >DroYak_CAF1 29959 87 + 1 ----GUUUACUACGAAAAUAUUUACAUUGCUGGCAUAAAAACUGCAUAAAUGUGAUUAUAUUUUCAUUGGCCUUUAUGCCUUAUGCCGU----CU ----.........(((((((((((((((....(((.......)))...)))))))..))))))))...(((......))).........----.. ( -17.40) >DroMoj_CAF1 25228 80 + 1 ----GUUUACAGCAAAAAUAUUUACACUGCAGGCAUAAAAACUGCAUAAAUGUGAUUAUAUUUUCAUUGGCUUUUAUGCCAA-----------CC ----..........((((((((((((.(((((.........)))))....)))))..)))))))..(((((......)))))-----------.. ( -16.50) >DroAna_CAF1 25526 84 + 1 GUGUGCUCGCAGCGAAAAUAUUUACAUUGCUGGCAUAAAAACUGCACAAAUGUGAUUAUAUUUUCAUUGGGUUUUAUGCCUU-----------CC (((((..(.(((.(((((((((((((((....(((.......)))...)))))))..)))))))).))).)...)))))...-----------.. ( -18.50) >consensus ____GUUUACAACGAAAAUAUUUACAUUGCUGGCAUAAAAACUGCAUAAAUGUGAUUAUAUUUUCAUUGGCUUUUAUGCCUU___________CC .............(((((((((((((((....(((.......)))...)))))))..))))))))...(((......)))............... (-14.10 = -13.97 + -0.14)


| Location | 12,215,447 – 12,215,538 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 81.27 |
| Mean single sequence MFE | -15.78 |
| Consensus MFE | -12.57 |
| Energy contribution | -13.13 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
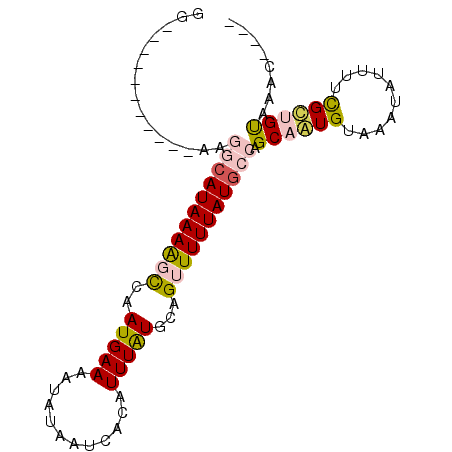
>3L_DroMel_CAF1 12215447 91 - 23771897 AAAACGACGGCAUAAGGCAUAAAGGCCAAUGAAAAUAUAAUCACAUUUAUGCAGUUUUUAUGCCAGCAAUGUAAAUAUUUUCGUAGUAAAC---- ...((((..((((..(((((((((((..(((((............)))))...)))))))))))....))))........)))).......---- ( -16.30) >DroVir_CAF1 32437 80 - 1 GG-----------CCAACAUAAAAGCCAAUGAAAAUAUAAUCACAUUUAUGCAGUUUUUAUGCCUGCAGUGUAAAUAUUUUCGCUGUAAAC---- ((-----------(..........)))...((((((((...(((.....(((((.........))))))))...)))))))).........---- ( -15.30) >DroWil_CAF1 34453 87 - 1 GA----UGAGUAUGAAAAAUAAAACUCAAUGAAAAUAUAAUCACAUUUAUGCAGUUUUUAUGCCAGCAAUGUAAAUAUUUUCGUUGUAAAC---- ..----.((((.(........).))))(((((((((((....(((((...(((.......)))....)))))..)))))))))))......---- ( -14.50) >DroYak_CAF1 29959 87 - 1 AG----ACGGCAUAAGGCAUAAAGGCCAAUGAAAAUAUAAUCACAUUUAUGCAGUUUUUAUGCCAGCAAUGUAAAUAUUUUCGUAGUAAAC---- ..----...((....(((......))).((((((((((....(((((...(((.......)))....)))))..)))))))))).))....---- ( -16.10) >DroMoj_CAF1 25228 80 - 1 GG-----------UUGGCAUAAAAGCCAAUGAAAAUAUAAUCACAUUUAUGCAGUUUUUAUGCCUGCAGUGUAAAUAUUUUUGCUGUAAAC---- ..-----------..(((((((((((..(((((............)))))...)))))))))))(((((((..........)))))))...---- ( -19.10) >DroAna_CAF1 25526 84 - 1 GG-----------AAGGCAUAAAACCCAAUGAAAAUAUAAUCACAUUUGUGCAGUUUUUAUGCCAGCAAUGUAAAUAUUUUCGCUGCGAGCACAC ..-----------..(((((((((....((....)).....(((....)))....))))))))).(((.((..........)).)))........ ( -13.40) >consensus GG___________AAGGCAUAAAAGCCAAUGAAAAUAUAAUCACAUUUAUGCAGUUUUUAUGCCAGCAAUGUAAAUAUUUUCGCUGUAAAC____ ...............(((((((((((..(((((............)))))...))))))))))).((((((..........))))))........ (-12.57 = -13.13 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:16 2006