
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,185,311 – 12,185,453 |
| Length | 142 |
| Max. P | 0.817889 |

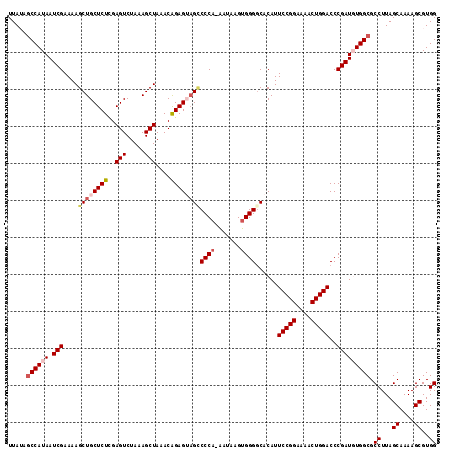
| Location | 12,185,311 – 12,185,427 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -28.52 |
| Energy contribution | -29.84 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12185311 116 + 23771897 UUAUAGCCAUAAUCGAAAAGCUGCUCUCGAGUCUAAAGCUAAACAGAGUUGCCCCAAAAUAAGUGGGGCACAUUCCGGAAAACUGGACCCGAUGUGGCGCCUUAGCAAAAGCGUGG .....((((((.(((.......(((((..(((.....)))....)))))(((((((.......)))))))...(((((....)))))..))))))))).((...((....))..)) ( -35.70) >DroSec_CAF1 19185 115 + 1 UUAUAACCAUAAUCGAAAAGCUGCUCUUGAGUCUAAAGCUAAACAGAGUAGUCCCA-AAUAAGUGGGGCACAUUCCGGAAAACUGGACCCGAUGUGGCGCCUUAGCAAAAGCGUGG ......(((((.(((......((((((..(((.....)))....))))))((((((-......))))))....(((((....)))))..))))))))((((((.....))).))). ( -30.80) >DroSim_CAF1 21610 115 + 1 UUAUAGCCAUAAUCGAAAAGCUGCUCUCGAGUCUAAAGCUAAACAGAGUAGCCCCA-AAUAAGUGGGGCACAUUCCGGAAAACUGGACCCGAUGUGGCGCCUUAGCAAAAGCGUGG .....((((((.(((......((((((..(((.....)))....))))))((((((-......))))))....(((((....)))))..))))))))).((...((....))..)) ( -37.40) >DroEre_CAF1 21152 113 + 1 UUA-AGCCAUAAUCGAAAAGCUGCUCUCGAGUCUAAAGCUAAACAGAGAAGACCCA-UAGAAGGGGGGCACAUUCCGGA-AUCUGGACCCGAUUUGGCGCCUUAGCAAAAGCGUGG ...-.((((.(((((....(((..((((.(((.....))).....))))...(((.-.....))).)))....(((((.-..)))))..))))))))).((...((....))..)) ( -29.30) >DroYak_CAF1 21770 113 + 1 UUA-AGCCAUAAUCGAAAAGCUGCUCUCGAGUCUAAAGCUAAACGGAGGAGCCCCA-AAGAAGUGGGCCACAUUCCGGA-AACUGGACCCGAUUUGGCGCCUUAGCAAAAGCGUGG ...-.((((.(((((....(((.((((..(((.....)))....)))).)))((((-......))))......((((..-..).)))..))))))))).((...((....))..)) ( -31.90) >consensus UUAUAGCCAUAAUCGAAAAGCUGCUCUCGAGUCUAAAGCUAAACAGAGUAGCCCCA_AAUAAGUGGGGCACAUUCCGGAAAACUGGACCCGAUGUGGCGCCUUAGCAAAAGCGUGG .....((((((.(((....((((((((..(((.....)))....))))))))((((.......))))......(((((....)))))..))))))))).((...((....))..)) (-28.52 = -29.84 + 1.32)


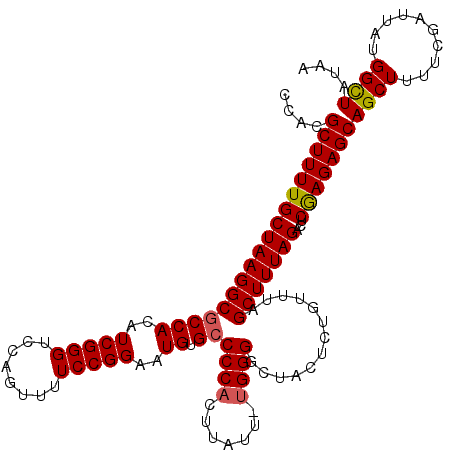
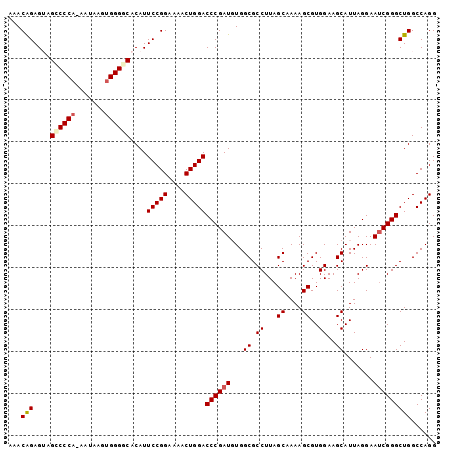
| Location | 12,185,311 – 12,185,427 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -34.44 |
| Consensus MFE | -28.28 |
| Energy contribution | -28.36 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12185311 116 - 23771897 CCACGCUUUUGCUAAGGCGCCACAUCGGGUCCAGUUUUCCGGAAUGUGCCCCACUUAUUUUGGGGCAACUCUGUUUAGCUUUAGACUCGAGAGCAGCUUUUCGAUUAUGGCUAUAA ....(((.(((..(((((((....(((((((.((((...((((...(((((((.......)))))))..))))...))))...)))))))..)).))))).)))....)))..... ( -38.60) >DroSec_CAF1 19185 115 - 1 CCACGCUUUUGCUAAGGCGCCACAUCGGGUCCAGUUUUCCGGAAUGUGCCCCACUUAUU-UGGGACUACUCUGUUUAGCUUUAGACUCAAGAGCAGCUUUUCGAUUAUGGUUAUAA ....((((((((((((((((((..(((((........)))))..)).))((((......-)))).............)))))))...)))))))...................... ( -29.60) >DroSim_CAF1 21610 115 - 1 CCACGCUUUUGCUAAGGCGCCACAUCGGGUCCAGUUUUCCGGAAUGUGCCCCACUUAUU-UGGGGCUACUCUGUUUAGCUUUAGACUCGAGAGCAGCUUUUCGAUUAUGGCUAUAA ....(((.(((..(((((((....(((((((.((((...((((.((.((((((......-)))))))).))))...))))...)))))))..)).))))).)))....)))..... ( -37.20) >DroEre_CAF1 21152 113 - 1 CCACGCUUUUGCUAAGGCGCCAAAUCGGGUCCAGAU-UCCGGAAUGUGCCCCCCUUCUA-UGGGUCUUCUCUGUUUAGCUUUAGACUCGAGAGCAGCUUUUCGAUUAUGGCU-UAA ....(((((....)))))((((((((((((((....-...)))...(((..(((.....-.)))...((((.(((((....)))))..)))))))....))))))).)))).-... ( -29.70) >DroYak_CAF1 21770 113 - 1 CCACGCUUUUGCUAAGGCGCCAAAUCGGGUCCAGUU-UCCGGAAUGUGGCCCACUUCUU-UGGGGCUCCUCCGUUUAGCUUUAGACUCGAGAGCAGCUUUUCGAUUAUGGCU-UAA ....(((((....)))))(((((((((((((.((((-..((((..(..((((.(.....-.)))))..)))))...))))...)))))(((((....))))))))).)))).-... ( -37.10) >consensus CCACGCUUUUGCUAAGGCGCCACAUCGGGUCCAGUUUUCCGGAAUGUGCCCCACUUAUU_UGGGGCUACUCUGUUUAGCUUUAGACUCGAGAGCAGCUUUUCGAUUAUGGCUAUAA ....((((((((((((((((((..(((((........)))))..)).))((((.......)))).............)))))))...)))))))((((..........)))).... (-28.28 = -28.36 + 0.08)

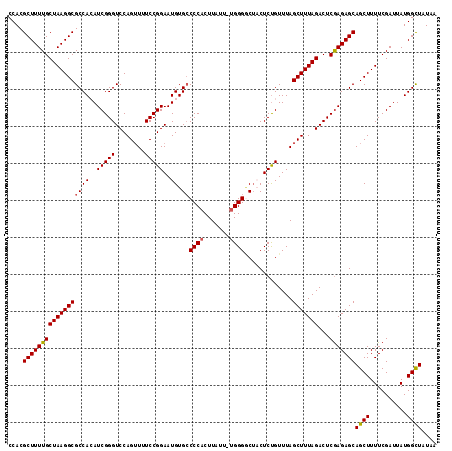
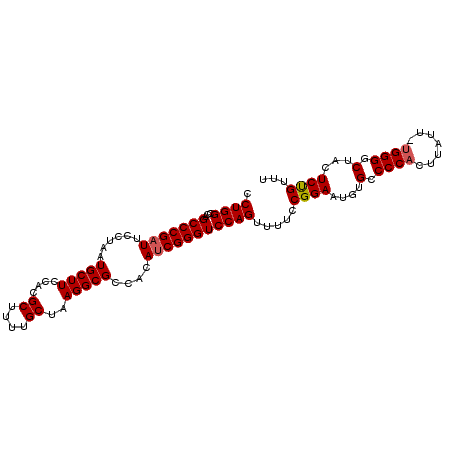
| Location | 12,185,351 – 12,185,453 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 93.68 |
| Mean single sequence MFE | -32.46 |
| Consensus MFE | -27.54 |
| Energy contribution | -28.10 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12185351 102 + 23771897 AAACAGAGUUGCCCCAAAAUAAGUGGGGCACAUUCCGGAAAACUGGACCCGAUGUGGCGCCUUAGCAAAAGCGUGGAAGCAUUAGGAAUCGGGCUGGCCAGG ...(.(((((((((((.......))))))).)))).).....((((.((((((...((.((...((....))..))..)).......))))))....)))). ( -35.20) >DroSec_CAF1 19225 101 + 1 AAACAGAGUAGUCCCA-AAUAAGUGGGGCACAUUCCGGAAAACUGGACCCGAUGUGGCGCCUUAGCAAAAGCGUGGAAGCAUUAGGAAUCGGGCUGGCCAGG ...(((.((.((((((-......))))))))..(((((....)))))((((((...((.((...((....))..))..)).......)))))))))...... ( -31.40) >DroSim_CAF1 21650 101 + 1 AAACAGAGUAGCCCCA-AAUAAGUGGGGCACAUUCCGGAAAACUGGACCCGAUGUGGCGCCUUAGCAAAAGCGUGGAAGCAUUAGGAAUCGGGCUGGCCAGG ...(((.((.((((((-......))))))))..(((((....)))))((((((...((.((...((....))..))..)).......)))))))))...... ( -34.10) >DroEre_CAF1 21191 100 + 1 AAACAGAGAAGACCCA-UAGAAGGGGGGCACAUUCCGGA-AUCUGGACCCGAUUUGGCGCCUUAGCAAAAGCGUGGAAGCAUUAGGAAUCGGGCUGGCCAGG ............(((.-.....))).(((.((.(((((.-..)))))((((((((.((.((...((....))..))..)).....)))))))).)))))... ( -29.60) >DroYak_CAF1 21809 100 + 1 AAACGGAGGAGCCCCA-AAGAAGUGGGCCACAUUCCGGA-AACUGGACCCGAUUUGGCGCCUUAGCAAAAGCGUGGAAGCAUUAGGAAGCGGGCUGGCCAGG ...(((((..(((((.-.....).))))....)))))..-..((((.(((..(((.((((.((....)).)))).)))((........)))))....)))). ( -32.00) >consensus AAACAGAGUAGCCCCA_AAUAAGUGGGGCACAUUCCGGAAAACUGGACCCGAUGUGGCGCCUUAGCAAAAGCGUGGAAGCAUUAGGAAUCGGGCUGGCCAGG ...(((....((((((.......))))))....(((((....)))))((((((...((.((...((....))..))..)).......)))))))))...... (-27.54 = -28.10 + 0.56)

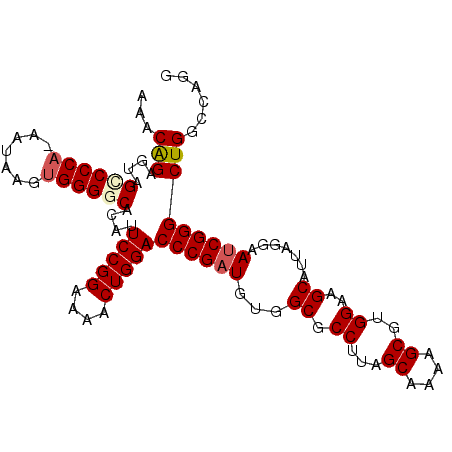
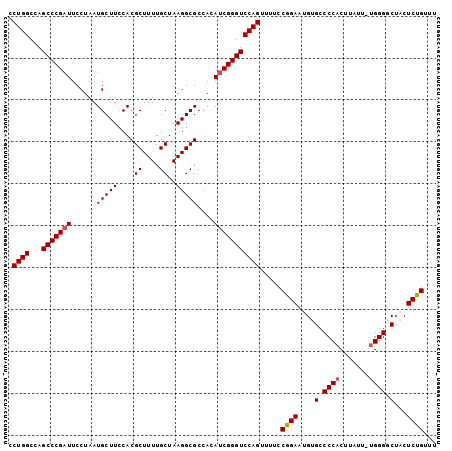
| Location | 12,185,351 – 12,185,453 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 93.68 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -24.48 |
| Energy contribution | -24.72 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12185351 102 - 23771897 CCUGGCCAGCCCGAUUCCUAAUGCUUCCACGCUUUUGCUAAGGCGCCACAUCGGGUCCAGUUUUCCGGAAUGUGCCCCACUUAUUUUGGGGCAACUCUGUUU .((((...(((((((......(((((....((....))..)))))....))))))))))).....((((...(((((((.......)))))))..))))... ( -34.40) >DroSec_CAF1 19225 101 - 1 CCUGGCCAGCCCGAUUCCUAAUGCUUCCACGCUUUUGCUAAGGCGCCACAUCGGGUCCAGUUUUCCGGAAUGUGCCCCACUUAUU-UGGGACUACUCUGUUU .((((...(((((((......(((((....((....))..)))))....))))))))))).....((((.((.(.((((......-)))).))).))))... ( -25.90) >DroSim_CAF1 21650 101 - 1 CCUGGCCAGCCCGAUUCCUAAUGCUUCCACGCUUUUGCUAAGGCGCCACAUCGGGUCCAGUUUUCCGGAAUGUGCCCCACUUAUU-UGGGGCUACUCUGUUU .((((...(((((((......(((((....((....))..)))))....))))))))))).....((((.((.((((((......-)))))))).))))... ( -33.00) >DroEre_CAF1 21191 100 - 1 CCUGGCCAGCCCGAUUCCUAAUGCUUCCACGCUUUUGCUAAGGCGCCAAAUCGGGUCCAGAU-UCCGGAAUGUGCCCCCCUUCUA-UGGGUCUUCUCUGUUU .((((...((((((((.....(((((....((....))..)))))...))))))))))))..-..((((..(.((((........-.)))).)..))))... ( -26.80) >DroYak_CAF1 21809 100 - 1 CCUGGCCAGCCCGCUUCCUAAUGCUUCCACGCUUUUGCUAAGGCGCCAAAUCGGGUCCAGUU-UCCGGAAUGUGGCCCACUUCUU-UGGGGCUCCUCCGUUU .((((...((((((........)).....((((((....)))))).......))))))))..-..((((..(..((((.(.....-.)))))..)))))... ( -31.40) >consensus CCUGGCCAGCCCGAUUCCUAAUGCUUCCACGCUUUUGCUAAGGCGCCACAUCGGGUCCAGUUUUCCGGAAUGUGCCCCACUUAUU_UGGGGCUACUCUGUUU .((((...(((((((......(((((....((....))..)))))....))))))))))).....((((....(.((((.......)))).)...))))... (-24.48 = -24.72 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:03 2006