| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,151,675 – 12,151,785 |
| Length | 110 |
| Max. P | 0.892004 |

| Location | 12,151,675 – 12,151,785 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 73.90 |
| Mean single sequence MFE | -21.50 |
| Consensus MFE | -9.38 |
| Energy contribution | -10.50 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12151675 110 + 23771897 AAUGACAUUUAUGUAUGUACAUAUG--UACAUACAUAUUUGCUCGCUUGACAUAUCACGGAGGUCGUUUUAAUACACGCCUUACCUGAUUUUUUUAACCCUCGGCUUGCCCA ...((((..(((((((((((....)--))))))))))..)).))..((((...((((..((((.(((........)))))))...))))....)))).....((....)).. ( -26.60) >DroSec_CAF1 28850 96 + 1 CA-GACAUUUAUGUACAAACAUACAUUUAUGUAC---------------AUAUAUCACGGAGGUCGUUUUAAUACACGCCUUACCUGAUUUUUUUAACCCUCGUUUGGCCCA ((-(((...((((((((............)))))---------------))).((((..((((.(((........)))))))...)))).............)))))..... ( -19.30) >DroSim_CAF1 29229 96 + 1 CA-GACAUUUAUGUACAUACAUACAUUUAUGUAC---------------AUAUAUCACGGAGGUCGUUUUAAUACACGCCUUACCUGAUUUUUUUAACCCUCGUUUUGCCCA .(-(((...((((((((((........)))))))---------------))).((((..((((.(((........)))))))...)))).............))))...... ( -19.20) >DroEre_CAF1 26327 86 + 1 CA-GACAUUUAUGUUCCC------------------------CCGCUUGACAUAUCACGGAGGUCGUUUUAAUACACGCCUCACCUGAUUUCUC-GAGCCUCGGCUUGCCCA ..-((((....))))...------------------------..((((((...((((.(((((.(((........))))))).).))))...))-))))...((....)).. ( -20.90) >consensus CA_GACAUUUAUGUACAUACAUACA__UAUGUAC_______________ACAUAUCACGGAGGUCGUUUUAAUACACGCCUUACCUGAUUUUUUUAACCCUCGGCUUGCCCA ............(((((((........)))))))...................((((..((((.(((........)))))))...))))....................... ( -9.38 = -10.50 + 1.12)

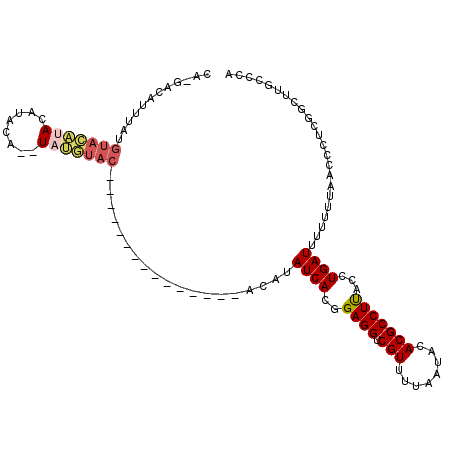
| Location | 12,151,675 – 12,151,785 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 73.90 |
| Mean single sequence MFE | -25.05 |
| Consensus MFE | -12.71 |
| Energy contribution | -14.27 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12151675 110 - 23771897 UGGGCAAGCCGAGGGUUAAAAAAAUCAGGUAAGGCGUGUAUUAAAACGACCUCCGUGAUAUGUCAAGCGAGCAAAUAUGUAUGUA--CAUAUGUACAUACAUAAAUGUCAUU ..((((((((...))))......((((((..((((((........))).))))).)))).))))....((.((..((((((((((--(....)))))))))))..))))... ( -30.00) >DroSec_CAF1 28850 96 - 1 UGGGCCAAACGAGGGUUAAAAAAAUCAGGUAAGGCGUGUAUUAAAACGACCUCCGUGAUAUAU---------------GUACAUAAAUGUAUGUUUGUACAUAAAUGUC-UG .((((...(((..((((.....(((((.(.....).)).))).....))))..)))....(((---------------(((((............))))))))...)))-). ( -19.90) >DroSim_CAF1 29229 96 - 1 UGGGCAAAACGAGGGUUAAAAAAAUCAGGUAAGGCGUGUAUUAAAACGACCUCCGUGAUAUAU---------------GUACAUAAAUGUAUGUAUGUACAUAAAUGUC-UG .(((((..(((..((((.....(((((.(.....).)).))).....))))..)))....(((---------------(((((((........))))))))))..))))-). ( -24.50) >DroEre_CAF1 26327 86 - 1 UGGGCAAGCCGAGGCUC-GAGAAAUCAGGUGAGGCGUGUAUUAAAACGACCUCCGUGAUAUGUCAAGCGG------------------------GGGAACAUAAAUGUC-UG .(((((.(((...((..-((....))..))..)))..............(((((((..........))))------------------------)))........))))-). ( -25.80) >consensus UGGGCAAAACGAGGGUUAAAAAAAUCAGGUAAGGCGUGUAUUAAAACGACCUCCGUGAUAUAU_______________GUACAUA__UGUAUGUAUGUACAUAAAUGUC_UG ..((((.................((((((..((((((........))).))))).))))...................(((((((........))))))).....))))... (-12.71 = -14.27 + 1.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:49 2006