
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,149,179 – 12,149,282 |
| Length | 103 |
| Max. P | 0.637692 |

| Location | 12,149,179 – 12,149,282 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.28 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -20.85 |
| Energy contribution | -21.93 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.637692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12149179 103 - 23771897 UUUACGUACUAUACAUAGAUAAAUUUUCCGGGUAAUCGGCGGAAAUUGAGU------------UAAAGUUUUGUGUCCGGUUGGCGCCAGUU---GGC--AAUUGGUGCUGGCCAACUUU ............((((((((((..((((((.........)))))).....)------------))....)))))))..(((..((((((((.---...--.))))))))..)))...... ( -28.30) >DroSec_CAF1 26380 103 - 1 UUUACGUACUAUACAUAUAUACAUUUUACGGGUAAUCGGCGGAAAUUGAGU------------UAAACUUUUGUGUCCGGUUGGCGCCAGUU---GGC--AAUUGGUGCUGGCCAACUUU ....((((..((..........))..))))(((.......(((....(((.------------....))).....)))(((..((((((((.---...--.))))))))..))).))).. ( -26.80) >DroSim_CAF1 26751 103 - 1 UUUACGUACUACACAUAUAUAAAUUUUCCGGGUAAUCGGCGGAAAUUGAGU------------UAAACUUUUGUGUCCGGUUGGCGCCAGUU---GGC--AAUUGGUGCUGGCCAACUUU ..........(((((.........((((((.........))))))..(((.------------....))).)))))..(((..((((((((.---...--.))))))))..)))...... ( -29.50) >DroEre_CAF1 23745 94 - 1 UUUACGUAU---------AUAACUUUUCCGGGUAAUCGGCGGAAAUUGACG------------UAAAGUUUUGUGUCCGGUUGGCGCCAUUU---GGC--CAUUGGUGCUGGCCAACUUU (((((((..---------......((((((.........))))))...)))------------))))...........(((..((((((...---...--...))))))..)))...... ( -29.10) >DroYak_CAF1 26660 99 - 1 UUUACGUAUU-------UUUAACUUUUCCGGAUAAUCGGCGGAAAUUGAGC------------UAAAGUUUUGUGUCCGGUUGGCGCCAGUUGGUGGC--CAUUGGUGCUGUCCAAGUUU ..........-------............(((((..(((((((....((((------------....))))....))).))))((((((((.((...)--))))))))))))))...... ( -26.40) >DroAna_CAF1 24462 107 - 1 -UUGCGAGC---------AUAACUUGGCCGGAUUUUCGCCCAUAAUCGAGUUGCAGUUAAGAUUAAAGUUUUGUGUCCGGUUGGCGCCACUU---GGCCAAAAUGGCGCUGGCCAACUUU -....(..(---------(((((((((.(((....))).)).(((((.............)))))))))..))))..)(((..((((((.((---.....)).))))))..)))...... ( -31.62) >consensus UUUACGUACU_______UAUAAAUUUUCCGGGUAAUCGGCGGAAAUUGAGU____________UAAAGUUUUGUGUCCGGUUGGCGCCAGUU___GGC__AAUUGGUGCUGGCCAACUUU ........................((((((.........)))))).................................(((..(((((((((........)))))))))..)))...... (-20.85 = -21.93 + 1.08)

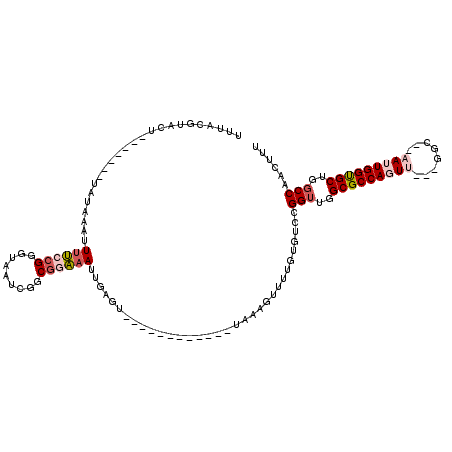
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:48 2006