
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
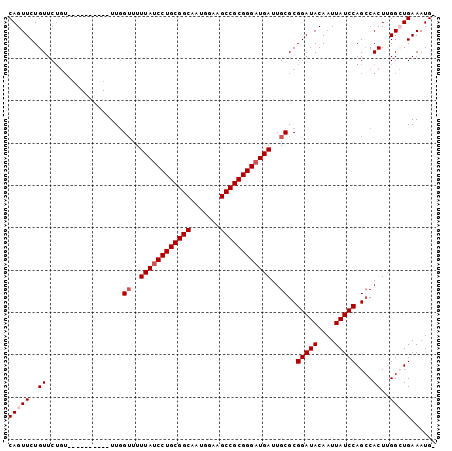
| Location | 12,145,983 – 12,146,104 |
| Length | 121 |
| Max. P | 0.981219 |

| Location | 12,145,983 – 12,146,084 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 87.44 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -28.25 |
| Energy contribution | -29.25 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
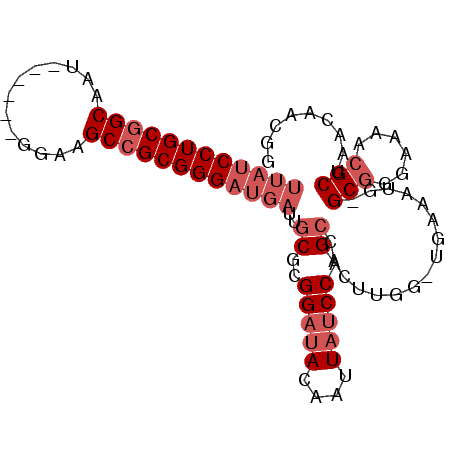
>3L_DroMel_CAF1 12145983 101 + 23771897 CAGUUCUGUUCUGUUCGUUGCAGUUUGGUUUUUAUCCUGCGGCAAUGGAAGCCGCGGGAUGAUUGCGCGGAUACAAUUAUCCAGCCACUCGGGUGAAAUGG ..(((((((.((((.....))))....((..((((((((((((.......))))))))))))..))))))).))..((((((........))))))..... ( -36.60) >DroSec_CAF1 23287 90 + 1 CAGUUCUGUUCUGU----------UUGGUUUUUAUCCUGCGGCAAUGGAAGCCGCGGGAUGAUUGCGCGGAUACAAUUAUCCAGGCACUUGGCUGAAAUG- (((((..(((((..----------...((..((((((((((((.......))))))))))))..))..(((((....)))))))).))..))))).....- ( -35.40) >DroSim_CAF1 23560 90 + 1 CAGUUCUGUUCUGU----------UUCGGUUUUAUCCUGCGGCAAUGGAAGCCGCGGGAUGAUUGCGCGGAUACAAUUAUCCAGCCACUUGGCUGAAAUG- ...........(((----------((((((.((((((((((((.......))))))))))))..)).)))).)))......(((((....))))).....- ( -35.50) >DroYak_CAF1 23305 94 + 1 CAGUUCUGUUC-----GUUGCUGUUUGGUUUUUAGCCUGCGGCAAUUGAAGCCGCGGGAUGAUUGCGCGGAUACAAUUAUCCAGCCACUUGG-UGAAAUG- ...........-----.(..(((..(((((.(((.((((((((.......)))))))).)))......(((((....))))))))))..)))-..)....- ( -29.70) >consensus CAGUUCUGUUCUGU__________UUGGUUUUUAUCCUGCGGCAAUGGAAGCCGCGGGAUGAUUGCGCGGAUACAAUUAUCCAGCCACUUGGCUGAAAUG_ (((((..((..................((..((((((((((((.......))))))))))))..))..(((((....)))))....))..)))))...... (-28.25 = -29.25 + 1.00)


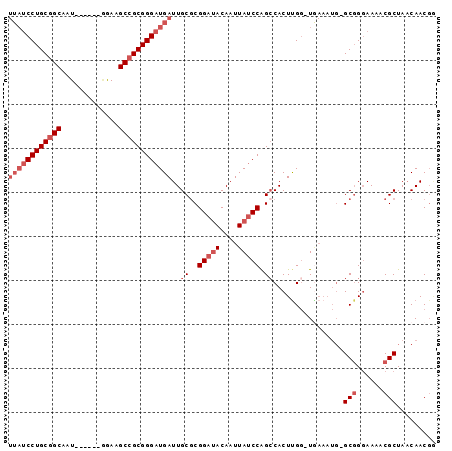
| Location | 12,145,983 – 12,146,084 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 87.44 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -22.25 |
| Energy contribution | -22.50 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12145983 101 - 23771897 CCAUUUCACCCGAGUGGCUGGAUAAUUGUAUCCGCGCAAUCAUCCCGCGGCUUCCAUUGCCGCAGGAUAAAAACCAAACUGCAACGAACAGAACAGAACUG .((.(((....((...((((((((....)))))).))..))((((.(((((.......))))).))))..........(((.......)))....))).)) ( -26.90) >DroSec_CAF1 23287 90 - 1 -CAUUUCAGCCAAGUGCCUGGAUAAUUGUAUCCGCGCAAUCAUCCCGCGGCUUCCAUUGCCGCAGGAUAAAAACCAA----------ACAGAACAGAACUG -((.(((......((((..(((((....)))))))))....((((.(((((.......))))).)))).........----------........))).)) ( -25.70) >DroSim_CAF1 23560 90 - 1 -CAUUUCAGCCAAGUGGCUGGAUAAUUGUAUCCGCGCAAUCAUCCCGCGGCUUCCAUUGCCGCAGGAUAAAACCGAA----------ACAGAACAGAACUG -...(((((((....)))))))..(((((......))))).((((.(((((.......))))).)))).........----------.............. ( -27.10) >DroYak_CAF1 23305 94 - 1 -CAUUUCA-CCAAGUGGCUGGAUAAUUGUAUCCGCGCAAUCAUCCCGCGGCUUCAAUUGCCGCAGGCUAAAAACCAAACAGCAAC-----GAACAGAACUG -((.(((.-....((.(((((((.(((((......))))).)))(((((((.......))))).))............)))).))-----.....))).)) ( -23.50) >consensus _CAUUUCAGCCAAGUGGCUGGAUAAUUGUAUCCGCGCAAUCAUCCCGCGGCUUCCAUUGCCGCAGGAUAAAAACCAA__________ACAGAACAGAACUG .((.(((......(((.(.(((((....)))))))))....((((.(((((.......))))).))))...........................))).)) (-22.25 = -22.50 + 0.25)


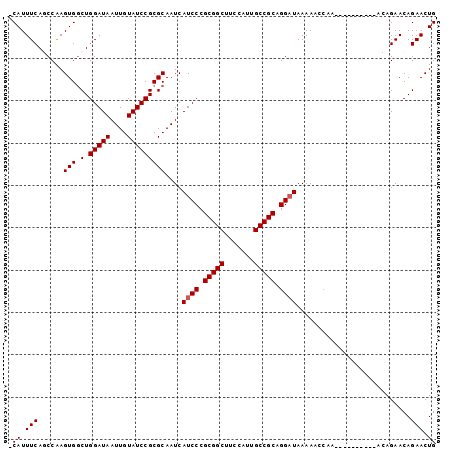
| Location | 12,146,014 – 12,146,104 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 86.11 |
| Mean single sequence MFE | -33.05 |
| Consensus MFE | -23.40 |
| Energy contribution | -24.90 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12146014 90 + 23771897 UUAUCCUGCGGCAAU------GGAAGCCGCGGGAUGAUUGCGCGGAUACAAUUAUCCAGCCACUCGGGUGAAAUGGGCGGGAAAACGCUA-CAACGG ((((((((((((...------....))))))))))))..((..(((((....))))).))(((....))).....((((......)))).-...... ( -34.50) >DroSec_CAF1 23308 90 + 1 UUAUCCUGCGGCAAU------GGAAGCCGCGGGAUGAUUGCGCGGAUACAAUUAUCCAGGCACUUGGCUGAAAUG-GCGGGAAAGCGCUAACAACGG ((((((((((((...------....))))))))))))..(((((((((....))))).....((((.(......)-.))))...))))......... ( -37.50) >DroSim_CAF1 23581 90 + 1 UUAUCCUGCGGCAAU------GGAAGCCGCGGGAUGAUUGCGCGGAUACAAUUAUCCAGCCACUUGGCUGAAAUG-GCGGGAAAACGCUAACAACGG ((((((((((((...------....))))))))))))......(((((....)))))((((....))))....((-(((......)))))....... ( -37.50) >DroEre_CAF1 20520 89 + 1 UUAUCCUGCGGCAAU------GAAAGCCGCGGGAUGAUUGCGCGGAUACAAUUAUCCAGCCACUUGG-UAAAAUG-GCGGCAAAACGCUAACAACGG ((((((((((((...------....))))))))))))((((..(((((....))))).((((.((..-..)).))-)).)))).............. ( -35.30) >DroYak_CAF1 23331 89 + 1 UUAGCCUGCGGCAAU------UGAAGCCGCGGGAUGAUUGCGCGGAUACAAUUAUCCAGCCACUUGG-UGAAAUG-GCGGGAAAACGCUAACAACUA ....((((((((...------....)))))))).....((.(((((((....))))).))))..(((-(....((-(((......)))))...)))) ( -30.60) >DroAna_CAF1 21478 86 + 1 ---UCCUGCCGCAAUUUUUGCAGAAGCCGCGGGAUGAUUGCUCGGAAACAAUUACCCGGCCACUUGC-U-------GCUGGAAAUCGCUAACAACGC ---.......((.(((((((((((((..(((((.((((((........))))))))).))..))).)-)-------)).)))))).))......... ( -22.90) >consensus UUAUCCUGCGGCAAU______GGAAGCCGCGGGAUGAUUGCGCGGAUACAAUUAUCCAGCCACUUGG_UGAAAUG_GCGGGAAAACGCUAACAACGG ((((((((((((.............))))))))))))..((..(((((....))))).))................(((......)))......... (-23.40 = -24.90 + 1.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:45 2006