
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,118,581 – 12,118,684 |
| Length | 103 |
| Max. P | 0.642135 |

| Location | 12,118,581 – 12,118,684 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.27 |
| Mean single sequence MFE | -30.57 |
| Consensus MFE | -20.06 |
| Energy contribution | -19.45 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
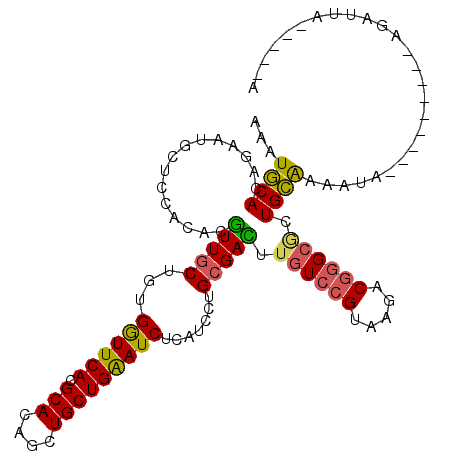
>3L_DroMel_CAF1 12118581 103 + 23771897 GGAUGCAGAGAAUGCUCCACUCGUUGCUGUGGUUCACGCACAGCUGCUGGAUCUCAUCCUGCGAUUUCUCCGUAAGGCGGGCGCUGCAAAAAUG------GAAGGUUA-----A (((((.(((......((((...((.(((((((....).)))))).))))))))))))))...(((((.(((((...((((...))))....)))------))))))).-----. ( -29.80) >DroPse_CAF1 1089 106 + 1 AAAUGCACAGAAUGCUCCAUACAUUGCUGUGGUUCACGCACAGCUGCUGAAUCUCCUCCUGCGAUUUGUCCGUAAGCCGGGCACUGCAAUUUA--------UAGAUUCAUAUUA ....(((...((((.......))))(((((((....).)))))))))(((((((.....((((...((((((.....)))))).)))).....--------.)))))))..... ( -27.70) >DroEre_CAF1 1083 109 + 1 GGAUGCAGAGAAUGCUCCACUCGUUGCUGUGGUUCACGCACAGCUGCUGGAUCUCAUCCUGCGACUUGUCCGUAAGGCGGGCGCUGCAAAAACAAAAACGGAAGGUUA-----A (((.(((.....))))))..((((((((((((....).))))))(((.((((...)))).(((.((((.((....))))))))).)))........))))).......-----. ( -35.30) >DroMoj_CAF1 1085 96 + 1 GAAUACACAAAAUGCUCCACACGUUGCUAUGAUUCACGCACAGCUGCUGAAUCUCAUCCUGCGACUUGUCCGUCAAACGUGCGCUGUGGA-------------GAACA-----A ..............(((((((((((((...((((((.(((....))))))))).......)))))..(..((.....))..)).))))))-------------)....-----. ( -27.60) >DroAna_CAF1 1084 101 + 1 AAAUGCACAGGAUGCUCCACACGUUGCUGUGGUUCACGCACAACUGCUGGAUCUCAUCCUGGGACUUGUCGGUCAGACGGGCCCUGCAGAAUUC--------AGGGAA-----A ...(((((((((((.((((((.((((.((((.....)))))))))).))))...)))))))((.((((((.....)))))).))))))......--------......-----. ( -37.50) >DroPer_CAF1 1089 106 + 1 AAAUGCACAGAAUGCUCCAGACAUUGCUGUGGUUCACGCACAGCUGCUGAAUCUCCUCCUGCGAUUUGUCCGUAAGCCGGGCACUGCAAUUUA--------UAGAUUGAUAUUA ....(((.....)))..(((.((..(((((((....).)))))))))))(((((.....((((...((((((.....)))))).)))).....--------.)))))....... ( -25.50) >consensus AAAUGCACAGAAUGCUCCACACGUUGCUGUGGUUCACGCACAGCUGCUGAAUCUCAUCCUGCGACUUGUCCGUAAGACGGGCGCUGCAAAAUA_________AGAUUA_____A ...((((...............(((((...((((((.(((....))))))))).......))))).((((((.....)))))).)))).......................... (-20.06 = -19.45 + -0.61)

| Location | 12,118,581 – 12,118,684 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.27 |
| Mean single sequence MFE | -31.20 |
| Consensus MFE | -22.44 |
| Energy contribution | -21.62 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12118581 103 - 23771897 U-----UAACCUUC------CAUUUUUGCAGCGCCCGCCUUACGGAGAAAUCGCAGGAUGAGAUCCAGCAGCUGUGCGUGAACCACAGCAACGAGUGGAGCAUUCUCUGCAUCC .-----......((------(((((.(((.(((((((.....))).)....))).((((...)))).)))((((((.(....)))))))...)))))))(((.....))).... ( -30.70) >DroPse_CAF1 1089 106 - 1 UAAUAUGAAUCUA--------UAAAUUGCAGUGCCCGGCUUACGGACAAAUCGCAGGAGGAGAUUCAGCAGCUGUGCGUGAACCACAGCAAUGUAUGGAGCAUUCUGUGCAUUU .....(((((((.--------....((((..((.(((.....))).))....))))....)))))))...((((((.(....)))))))(((((((((......))))))))). ( -35.70) >DroEre_CAF1 1083 109 - 1 U-----UAACCUUCCGUUUUUGUUUUUGCAGCGCCCGCCUUACGGACAAGUCGCAGGAUGAGAUCCAGCAGCUGUGCGUGAACCACAGCAACGAGUGGAGCAUUCUCUGCAUCC .-----........((((........(((.(((((((.....)))....).))).((((...)))).)))((((((.(....))))))))))).(..(((....)))..).... ( -29.70) >DroMoj_CAF1 1085 96 - 1 U-----UGUUC-------------UCCACAGCGCACGUUUGACGGACAAGUCGCAGGAUGAGAUUCAGCAGCUGUGCGUGAAUCAUAGCAACGUGUGGAGCAUUUUGUGUAUUC .-----(((((-------------..(((...((.((((((((......))))...)))).(((((((((....))).))))))...))...)))..)))))............ ( -26.00) >DroAna_CAF1 1084 101 - 1 U-----UUCCCU--------GAAUUCUGCAGGGCCCGUCUGACCGACAAGUCCCAGGAUGAGAUCCAGCAGUUGUGCGUGAACCACAGCAACGUGUGGAGCAUCCUGUGCAUUU .-----......--------......((((((((..(((.....)))..))))(((((((...((((.(((((((..(((...))).))))).)))))).)))))))))))... ( -35.20) >DroPer_CAF1 1089 106 - 1 UAAUAUCAAUCUA--------UAAAUUGCAGUGCCCGGCUUACGGACAAAUCGCAGGAGGAGAUUCAGCAGCUGUGCGUGAACCACAGCAAUGUCUGGAGCAUUCUGUGCAUUU .............--------.....(((..((.(((.....))).))....((((((.(...(((((((((((((.(....)))))))..)).))))).).)))))))))... ( -29.90) >consensus U_____UAACCU_________AAUUUUGCAGCGCCCGCCUUACGGACAAAUCGCAGGAUGAGAUCCAGCAGCUGUGCGUGAACCACAGCAACGUGUGGAGCAUUCUGUGCAUUC ..........................(((((((.(((.....)))......)))((((((...((((...((((((.(....)))))))......)))).)))))).))))... (-22.44 = -21.62 + -0.83)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:33 2006