| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,112,242 – 12,112,348 |
| Length | 106 |
| Max. P | 0.571094 |

| Location | 12,112,242 – 12,112,348 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.28 |
| Mean single sequence MFE | -41.65 |
| Consensus MFE | -25.44 |
| Energy contribution | -26.30 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.571094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12112242 106 - 23771897 AUGGUCGGUUGGAGCCGUUUGCUGUCGCCGGCGGCGAAGUUCGCCAGCUACCGGGCUGUCCUGCAAGAGCCCGCGUGCCGGAAUGGUGAGCAUUUCUUGAAA------AUAU- ((..(((((....)))(((..((((..(((((((((.....))))....((((((((.(......).)))))).))))))).))))..))).......))..------))..- ( -41.80) >DroPse_CAF1 8264 105 - 1 AUGGUCGGUUGGAGCCGUUUGCUGUCGCCAGCGGCGCAAUUCGCCAACUACCGCGCCGUUCUGCAAGCGCCGGCUUGCCGGAAUGGUAAGUAGAGCUUAGA-------UCAG- .((((..((((..((((((.((....)).)))))).))))..)))).((((...((((((((((((((....))))).)))))))))..))))........-------....- ( -45.10) >DroEre_CAF1 6097 105 - 1 AUGGUCGGUUGGAGCCGUUUGCUUUCGCCGGCGGCGAAGUUCGCCAGUUACCGGGCUGUCCUGCAGGAGCCCGCUUGCCGGAAUGGUGAGCACUCCUUGGA-------UUAU- ((((((.(..((((..(((..((....(((((((((.....))))......(((((..(((....))))))))...)))))...))..))).)))).).))-------))))- ( -47.20) >DroMoj_CAF1 6058 113 - 1 AUGGUUGGUUGGAGCCGUUUGCUGCUGCCAGCGGCACAAUUCGCCAAGAAUAGGGCUGUACUGCUGACACCAGCCACAAGGAAUGGUAAGCACAACUGCUAAUGCAUCAGAUG .((((..((((..((((((.((....)).)))))).))))..)))).......(((.((..((((...((((.((....))..)))).))))..)).)))............. ( -35.70) >DroAna_CAF1 5934 103 - 1 AUGGUCGGUUGGAGCCGUAUGCUGUCGCCGGCGGCAUCUUUCGCGAAGUACCGCGCCGUGCUUCAGACACCUGCCUGCCGAAAUGGUAAGUAUCGCCAGUAA------A---- .(((.((((....(((((..((....))((((((((....((..(((((((......))))))).))....)))).))))..)))))....)))))))....------.---- ( -35.00) >DroPer_CAF1 6135 105 - 1 AUGGUCGGUUGGAGCCGUUUGCUGUCGCCAGCGGCGCAAUUCGCCAACUACCGCGCCGUUCUGCAAGCGCCGGCUUGCCGGAAUGGUAAGUAGAGCUUAGA-------UCAG- .((((..((((..((((((.((....)).)))))).))))..)))).((((...((((((((((((((....))))).)))))))))..))))........-------....- ( -45.10) >consensus AUGGUCGGUUGGAGCCGUUUGCUGUCGCCAGCGGCGCAAUUCGCCAACUACCGCGCCGUCCUGCAAGCGCCCGCUUGCCGGAAUGGUAAGCACAGCUUGGA_______ACAU_ .(((.(((.....((((((.((....)).)))))).....))))))..(((((..(((....((((((....)))))))))..)))))......................... (-25.44 = -26.30 + 0.86)

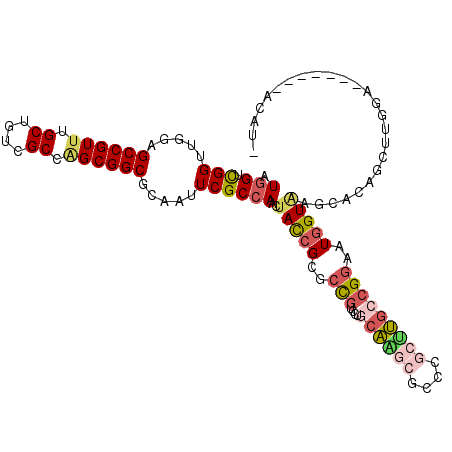

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:31 2006