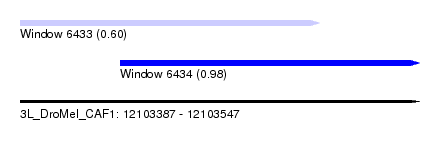
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,103,387 – 12,103,547 |
| Length | 160 |
| Max. P | 0.979943 |

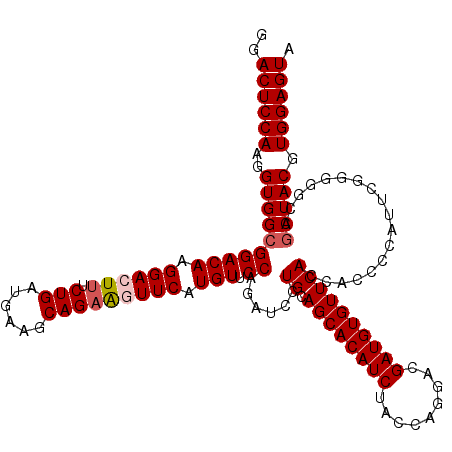
| Location | 12,103,387 – 12,103,507 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -43.62 |
| Consensus MFE | -40.88 |
| Energy contribution | -41.28 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601116 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12103387 120 + 23771897 CAGUGGCAUUGCUGGCUACUCUCGGCCAGGCCAAGCACCUGGACUCCAAGGUGGCGGACAAGGACUUUCUGAUGAAACAGAAGUUCAUGUACCAGAUCCUGCAGCACAUCUACCAGGACG ..((((...((((((((((.((.(((((((.......)))))...)).)))))))(((((.((((((.(((......))))))))).))).))........)))))...))))....... ( -40.80) >DroSec_CAF1 4510 120 + 1 CGGUGGCAUUGCUGGCUACUCUCGGCCAGGCCAAGCACCUGGACUCCAAGGUGGCGGACAAGGACUUUCUGAUGAAGCAGAAGUUCGUGUUCCACAUCCUGCAGCACAUCUACCAGGCCG .(((((...((((((((((.((.(((((((.......)))))...)).)))))))(((((.((((((.(((......))))))))).))))).........)))))...)))))...... ( -45.30) >DroSim_CAF1 3890 120 + 1 CGGUGGCAUUGCUGGCUACUCUCGGCCAGGCCAAGCACCUGGACUCCAAGGUGGCGGACAAGGACUUUCUGGUGAAGCAGCAGUUCAUGUUCCACAUCCUGCAGCACAUCUACCAGGAUG .(((((...((((((((((.((.(((((((.......)))))...)).)))))))(((((.(((((..(((......))).))))).))))).........)))))...)))))...... ( -42.90) >DroEre_CAF1 4027 120 + 1 CGGUGGCAUUGCUGGCUGCUCUGGGCCAGGCCAAGCACCUGGACUCCAAGGUGGCGGACAAGGACUUCCUGACCAAGCAGAGCUUCAUGUUCCAGAUCCUGCAGCACAUCUACCAGGACG .(((((...(((((((..(..(((((((((.......))))).).)))..)..))((((.(((....))).........((((.....))))..).)))..)))))...)))))...... ( -43.50) >DroYak_CAF1 4028 120 + 1 CGGUGGCAUUGCUGGCUGCUCUGGGCCAGGCCAAGCACCUGGACUCCAAGGUGGCGGACAAGGACUUUCUGAUGAAGCAGAACUUCGUGUUCCAGAUCCUGCAGCACAUCUACCAGGACG .(((((...(((((((..(..(((((((((.......))))).).)))..)..)).....((((...((((((((((.....))))))....)))))))).)))))...)))))...... ( -45.60) >consensus CGGUGGCAUUGCUGGCUACUCUCGGCCAGGCCAAGCACCUGGACUCCAAGGUGGCGGACAAGGACUUUCUGAUGAAGCAGAAGUUCAUGUUCCAGAUCCUGCAGCACAUCUACCAGGACG .(((((...((((((((((.((.(((((((.......)))))...)).)))))))(((((.((((((.(((......))))))))).))).))........)))))...)))))...... (-40.88 = -41.28 + 0.40)

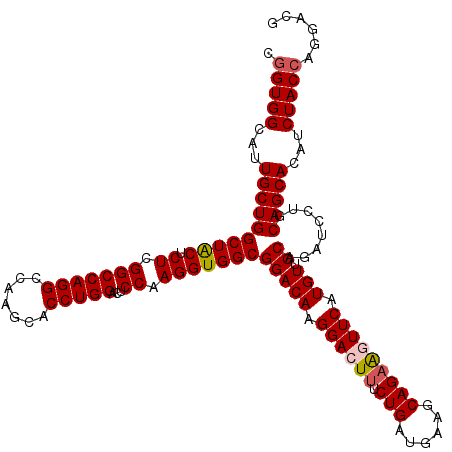
| Location | 12,103,427 – 12,103,547 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.67 |
| Mean single sequence MFE | -44.32 |
| Consensus MFE | -35.40 |
| Energy contribution | -36.04 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12103427 120 + 23771897 GGACUCCAAGGUGGCGGACAAGGACUUUCUGAUGAAACAGAAGUUCAUGUACCAGAUCCUGCAGCACAUCUACCAGGACGAUGUGUUCACCACGCCCUUCGGGGGCAGCUACGUGGAGUA ..((((((..((((((((((.((((((.(((......))))))))).))).))......(((((((((((.........)))))))).......(((....))))))))))).)))))). ( -48.90) >DroSec_CAF1 4550 120 + 1 GGACUCCAAGGUGGCGGACAAGGACUUUCUGAUGAAGCAGAAGUUCGUGUUCCACAUCCUGCAGCACAUCUACCAGGCCGAUGUGUUCACCACCCCAUUCGGAGGCAGCUACGUGGAGUA ..((((((..((((((((((.((((((.(((......))))))))).))))).......(((((((((((.........)))))))).....(.((....)).))))))))).)))))). ( -42.50) >DroSim_CAF1 3930 120 + 1 GGACUCCAAGGUGGCGGACAAGGACUUUCUGGUGAAGCAGCAGUUCAUGUUCCACAUCCUGCAGCACAUCUACCAGGAUGAUGUGUUCACCACCCCAUUCGGGGGCAGCUACGUGGAGUA ..((((((..((((((((((.(((((..(((......))).))))).))))).......(((((((((((.........)))))))).....((((....)))))))))))).)))))). ( -47.60) >DroEre_CAF1 4067 120 + 1 GGACUCCAAGGUGGCGGACAAGGACUUCCUGACCAAGCAGAGCUUCAUGUUCCAGAUCCUGCAGCACAUCUACCAGGACGAUGUGUUCACAACCCACUUCCCGGCCACCUACAUGGAGUA ..(((((((((((((((...(((....)))..))..((((.(..((........)).)))))((((((((.........))))))))................)))))))...)))))). ( -40.00) >DroYak_CAF1 4068 120 + 1 GGACUCCAAGGUGGCGGACAAGGACUUUCUGAUGAAGCAGAACUUCGUGUUCCAGAUCCUGCAGCACAUCUACCAGGACGAUGUGUUCACAACCCCAUUCUCCGCCAGCUACGUGGAGUA ..((((((((.(((((((..((((...((((((((((.....))))))....))))))))(.((((((((.........)))))))))............))))))).))...)))))). ( -42.60) >consensus GGACUCCAAGGUGGCGGACAAGGACUUUCUGAUGAAGCAGAAGUUCAUGUUCCAGAUCCUGCAGCACAUCUACCAGGACGAUGUGUUCACCACCCCAUUCGGGGGCAGCUACGUGGAGUA ..((((((..((((((((((.((((((.(((......))))))))).))).))......((.((((((((.........))))))))))..................))))).)))))). (-35.40 = -36.04 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:23 2006