| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,098,964 – 12,099,074 |
| Length | 110 |
| Max. P | 0.959776 |

| Location | 12,098,964 – 12,099,074 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.45 |
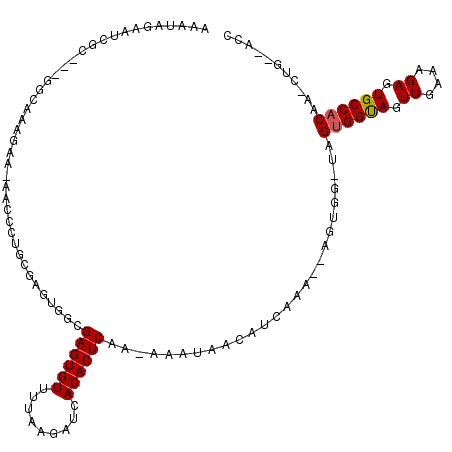
| Mean single sequence MFE | -24.72 |
| Consensus MFE | -17.69 |
| Energy contribution | -17.72 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
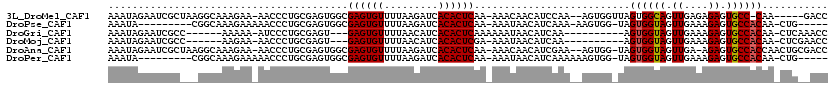
>3L_DroMel_CAF1 12098964 110 - 23771897 AAAUAGAAUCGCUAAGGCAAAGAA-AACCCUGCGAGUGGCGAGUGUUUUAAGAUCACACUCAA-AAACAACAUCCAA--AGUGGUUAGUGGCAGUUGAGAGAGUGCC-CAA-----GACC ..........((((..(((.....-.....)))...))))((((((.........))))))..-.............--...((((...((((.((....)).))))-...-----)))) ( -25.10) >DroPse_CAF1 5526 102 - 1 AAAUA---------CGGCAAAGAAAAACCCUGCGAGUGGCGAGUGUUUUAAGAUCACACUCAA-AAAUAACAUCAAA-AAGUGG-UAGUGGUAGUUGAAAGAGUGCCACAA-CUG----- ...((---------(.((.............((.....))((((((.........))))))..-.............-..)).)-))((((((.((....)).))))))..-...----- ( -24.60) >DroGri_CAF1 31005 99 - 1 AAAUAGAAUCGCC------AAAAA-AUCCCUGCGAGU---GAGUGUUUUAACAUCACACUCAAAAAAUAACAUCAA----------AGUGGUAGUUGAAAGAGUGCCACAA-CUCAAACC .............------.....-........((((---((((((.........))))))...............----------.((((((.((....)).)))))).)-)))..... ( -22.90) >DroMoj_CAF1 32043 98 - 1 AAAUAGAAUCGCC------AAGAA-AACCCUGCGAGU---GAGUGUUUUAACAUCACACUCGA-AAAUAACAUCAA----------AGUGGUAGUUGAAAGAGUGCCACAA-CUCGAACC .............------.....-.......(((((---((((((.........))))))..-............----------.((((((.((....)).)))))).)-)))).... ( -24.60) >DroAna_CAF1 7803 114 - 1 AAAUAGAAUCGCUAAGGCAAAGAA-AACCCUGCGAGUGGCGAGUGUUUUAAGAUCACACUCAA-AAACAACAUCGAA--AGUGG-UAGUGGUAGUUGA-AGAGUGCCACCAACUGCGACC ........((((...((((..((.-.((((((((((((..((...........)).)))))..-......((.(...--.))))-))).)))..))..-....)))).......)))).. ( -26.50) >DroPer_CAF1 5509 103 - 1 AAAUA---------CGGCAAAGAAAAACCCUGCGAGUGGCGAGUGUUUUAAGAUCACACUCAA-AAAUAACAUCAAAAAAGUGG-UAGUGGUAGUUGAAAGAGUGCCACAA-CUG----- ...((---------(.((.............((.....))((((((.........))))))..-................)).)-))((((((.((....)).))))))..-...----- ( -24.60) >consensus AAAUAGAAUCGC___GGCAAAGAA_AACCCUGCGAGUGGCGAGUGUUUUAAGAUCACACUCAA_AAAUAACAUCAAA__AGUGG_UAGUGGUAGUUGAAAGAGUGCCACAA_CUG__ACC ........................................((((((.........))))))..........................((((((.((....)).))))))........... (-17.69 = -17.72 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:18 2006